Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum
- PMID: 10725325
- PMCID: PMC2174309
- DOI: 10.1083/jcb.148.6.1123
Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum
Abstract
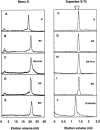
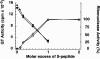
Newly synthesized glycoproteins interact during folding and quality control in the ER with calnexin and calreticulin, two lectins specific for monoglucosylated oligosaccharides. Binding and release are regulated by two enzymes, glucosidase II and UDP-Glc:glycoprotein:glycosyltransferase (GT), which cyclically remove and reattach the essential glucose residues on the N-linked oligosaccharides. GT acts as a folding sensor in the cycle, selectively reglucosylating incompletely folded glycoproteins and promoting binding of its substrates to the lectins. To investigate how nonnative protein conformations are recognized and directed to this unique chaperone system, we analyzed the interaction of GT with a series of model substrates with well defined conformations derived from RNaseB. We found that conformations with slight perturbations were not reglucosylated by GT. In contrast, a partially structured nonnative form was efficiently recognized by the enzyme. When this form was converted back to a nativelike state, concomitant loss of recognition by GT occurred, reproducing the reglucosylation conditions observed in vivo with isolated components. Moreover, fully unfolded conformers were poorly recognized. The results indicated that GT is able to distinguish between different nonnative conformations with a distinct preference for partially structured conformers. The findings suggest that discrete populations of nonnative conformations are selectively reglucosylated to participate in the calnexin/calreticulin chaperone pathway.
Figures







Similar articles
-
Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation.Biochem J. 2000 May 15;348 Pt 1(Pt 1):1-13. Biochem J. 2000. PMID: 10794707 Free PMC article. Review.
-
Protein glucosylation and its role in protein folding.Annu Rev Biochem. 2000;69:69-93. doi: 10.1146/annurev.biochem.69.1.69. Annu Rev Biochem. 2000. PMID: 10966453 Review.
-
Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT.EMBO J. 2005 May 4;24(9):1730-8. doi: 10.1038/sj.emboj.7600645. Epub 2005 Apr 14. EMBO J. 2005. PMID: 15861139 Free PMC article.
-
Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells.Biochim Biophys Acta. 1999 Jan 6;1426(2):287-95. doi: 10.1016/s0304-4165(98)00130-5. Biochim Biophys Acta. 1999. PMID: 9878790 Review.
-
Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase.Nat Struct Biol. 2000 Apr;7(4):278-80. doi: 10.1038/74035. Nat Struct Biol. 2000. PMID: 10742170
Cited by
-
UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates.Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):86-91. doi: 10.1073/pnas.262661199. Epub 2002 Dec 23. Proc Natl Acad Sci U S A. 2003. PMID: 12518055 Free PMC article.
-
Defining the glycan destruction signal for endoplasmic reticulum-associated degradation.Mol Cell. 2008 Dec 26;32(6):870-7. doi: 10.1016/j.molcel.2008.11.017. Mol Cell. 2008. PMID: 19111666 Free PMC article.
-
Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals.Diabetes. 2008 Sep;57(9):2438-44. doi: 10.2337/db08-0604. Epub 2008 Jun 20. Diabetes. 2008. PMID: 18567819 Free PMC article.
-
Endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins and mutant P23H rhodopsin in photoreceptor cells.Adv Exp Med Biol. 2012;723:559-65. doi: 10.1007/978-1-4614-0631-0_71. Adv Exp Med Biol. 2012. PMID: 22183378 Free PMC article. Review. No abstract available.
-
Oligomannose-Type Glycan Processing in the Endoplasmic Reticulum and Its Importance in Misfolding Diseases.Biology (Basel). 2022 Jan 27;11(2):199. doi: 10.3390/biology11020199. Biology (Basel). 2022. PMID: 35205066 Free PMC article. Review.
References
-
- Allende J.E., Richards F.M. The action of trypsin of ribonuclease-S. Biochemistry. 1962;1:295–304. - PubMed
-
- Bergeron J.J., Brenner M.B., Thomas D.Y., Williams D.B. Calnexina membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. - PubMed
-
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. - PubMed
-
- Cannon K., Helenuis A. Trimming and readdition of glucose to N-linked oligosaccharides determines calnexin association of a substrate glycoprotein in living cells. J. Biol. Chem. 1999;274:7537–7544. - PubMed
-
- Ellgard L., Molinari M., Helenius A. Setting the standardsquality control in the secretory pathway. Science. 1999;286:1882–1888. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

