Ubinuclein, a novel nuclear protein interacting with cellular and viral transcription factors
- PMID: 10725330
- PMCID: PMC2174308
- DOI: 10.1083/jcb.148.6.1165
Ubinuclein, a novel nuclear protein interacting with cellular and viral transcription factors
Abstract
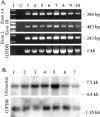
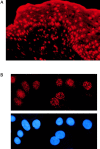
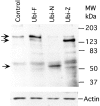
The major target tissues for Epstein-Barr virus (EBV) infection are B lymphocytes and epithelial cells of the oropharyngeal zone. The product of the EBV BZLF1 early gene, EB1, a member of the basic leucine-zipper family of transcription factors, interacts with both viral and cellular promoters and transcription factors, modulating the reactivation of latent EBV infection. Here, we characterize a novel cellular protein interacting with the basic domains of EB1 and c-Jun, and competing of their binding to the AP1 consensus site. The transcript is present in a wide variety of human adult, fetal, and tumor tissues, and the protein is detected in the nuclei throughout the human epidermis and as either grainy or punctuate nuclear staining in the cultured keratinocytes. The overexpression of tagged cDNA constructs in keratinocytes revealed that the NH(2) terminus is essential for the nuclear localization, while the central domain is responsible for the interaction with EB1 and for the phenotype of transfected keratinocytes similar to terminal differentiation. The gene was identified in tail-to-tail orientation with the periplakin gene (PPL) in human chromosome 16p13.3 and in a syntenic region in mouse chromosome 16. We designated this novel ubiquitously expressed nuclear protein as ubinuclein and the corresponding gene as UBN1.
Figures




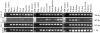



References
-
- Aho S., McLean W.H.I., Li K., Uitto J. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin gene. Genomics. 1998;48:242–247. - PubMed
-
- Aho S., Rothenberger K., Tan E.M.L., Ryoo Y.W., Cho B.H., McLean W.H.I., Uitto J. Human periplakingenomic organization in a clonally unstable region of chromosome 16p with an abundance of repetitive sequence elements. Genomics. 1999;56:160–168. - PubMed
-
- Aït-Ahmed O., Thomas-Cavallin M., Rosset R. Isolation and characterization of the Drosophila genome which contains a cluster of differentially expressed maternal genes (yema gene region) Dev. Biol. 1987;122:153–162. - PubMed
-
- Aït-Ahmed O., Bellon B., Capri M., Joblet C., Thomas-Delaage M. The yemanuclein-αa new Drosophila DNA binding protein specific for the oocyte nucleus. Mech. Develop. 1992;37:69–80. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

