Toc64, a new component of the protein translocon of chloroplasts
- PMID: 10725334
- PMCID: PMC2174300
- DOI: 10.1083/jcb.148.6.1213
Toc64, a new component of the protein translocon of chloroplasts
Abstract
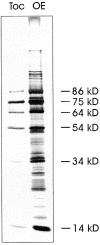
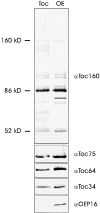
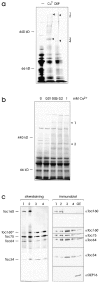
A subunit of the preprotein translocon of the outer envelope of chloroplasts (Toc complex) of 64 kD is described, Toc64. Toc64 copurifies on sucrose density gradients with the isolated Toc complex. Furthermore, it can be cross-linked in intact chloroplasts to a high molecular weight complex containing both Toc and Tic subunits and a precursor protein. The 0 A cross-linker CuCl(2) yields the reversible formation of disulfide bridge(s) between Toc64 and the established Toc complex subunits in purified outer envelope membranes. Toc64 contains three tetratricopeptide repeat motifs that are exposed at the chloroplast cytosol interface. We propose that Toc64 functions early in preprotein translocation, maybe as a docking protein for cytosolic cofactors of the protein import into chloroplasts.
Figures


Similar articles
-
Toc64--a preprotein-receptor at the outer membrane with bipartide function.J Mol Biol. 2007 Apr 13;367(5):1330-46. doi: 10.1016/j.jmb.2007.01.047. Epub 2007 Jan 24. J Mol Biol. 2007. PMID: 17306301
-
Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts.Mol Biol Cell. 2004 Nov;15(11):5130-44. doi: 10.1091/mbc.e04-05-0405. Epub 2004 Aug 18. Mol Biol Cell. 2004. PMID: 15317846 Free PMC article.
-
A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide.FEBS Lett. 1998 Dec 11;441(1):59-62. doi: 10.1016/s0014-5793(98)01525-7. FEBS Lett. 1998. PMID: 9877165
-
Toc, tic, and chloroplast protein import.Biochim Biophys Acta. 2002 Jun 12;1590(1-3):177-89. doi: 10.1016/s0167-4889(02)00176-3. Biochim Biophys Acta. 2002. PMID: 12180471 Review.
-
Toc, Tic, and chloroplast protein import.Biochim Biophys Acta. 2001 Dec 12;1541(1-2):64-79. doi: 10.1016/s0167-4889(01)00147-1. Biochim Biophys Acta. 2001. Corrected and republished in: Biochim Biophys Acta. 2002 Jun 12;1590(1-3):177-89. doi: 10.1016/s0167-4889(02)00176-3. PMID: 11750663 Corrected and republished. Review.
Cited by
-
14-3-3 proteins and plant development.Plant Mol Biol. 2002 Dec;50(6):1019-29. doi: 10.1023/a:1021295604109. Plant Mol Biol. 2002. PMID: 12516869
-
Chaperone receptors: guiding proteins to intracellular compartments.Protoplasma. 2012 Jan;249(1):21-30. doi: 10.1007/s00709-011-0270-9. Epub 2011 Apr 3. Protoplasma. 2012. PMID: 21461941 Review.
-
Molecular characterization and expression analysis of chloroplast protein import components in tomato (Solanum lycopersicum).PLoS One. 2014 Apr 21;9(4):e95088. doi: 10.1371/journal.pone.0095088. eCollection 2014. PLoS One. 2014. PMID: 24751891 Free PMC article.
-
A GTP-driven motor moves proteins across the outer envelope of chloroplasts.Proc Natl Acad Sci U S A. 2003 Apr 15;100(8):4604-9. doi: 10.1073/pnas.0730860100. Epub 2003 Mar 28. Proc Natl Acad Sci U S A. 2003. PMID: 12665619 Free PMC article.
-
The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides.Biophys J. 2002 Aug;83(2):899-911. doi: 10.1016/S0006-3495(02)75216-8. Biophys J. 2002. PMID: 12124272 Free PMC article.
References
-
- Anderson C.W., Straus J.W., Dudock B.S. Preparation of a cell-free protein-synthesizing system from wheat germ. Methods Enzymol. 1983;101:635–644. - PubMed
-
- Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. - PubMed
-
- Bölter B., May T., Soll J. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytical fragment of a larger polypeptide. FEBS Lett. 1998;441:59–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

