The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3
- PMID: 10725350
- PMCID: PMC16309
- DOI: 10.1073/pnas.97.7.3735
The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3
Abstract
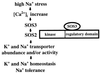
The Arabidopsis thaliana SOS2 and SOS3 genes are required for intracellular Na(+) and K(+) homeostasis and plant tolerance to high Na(+) and low K(+) environments. SOS3 is an EF hand type calcium-binding protein having sequence similarities with animal neuronal calcium sensors and the yeast calcineurin B. SOS2 is a serine/threonine protein kinase in the SNF1/AMPK family. We report here that SOS3 physically interacts with and activates SOS2 protein kinase. Genetically, sos2sos3 double mutant analysis indicates that SOS2 and SOS3 function in the same pathway. Biochemically, SOS2 interacts with SOS3 in the yeast two-hybrid system and in vitro binding assays. The interaction is mediated by the C-terminal regulatory domain of SOS2. SOS3 activates SOS2 protein kinase activity in a Ca(2+)-dependent manner. Therefore, SOS3 and SOS2 define a novel regulatory pathway important for the control of intracellular ion homeostasis and salt tolerance in plants.
Figures





References
-
- Stein W D. Channels, Carriers and Pumps: An Introduction to Membrane Transport. New York: Academic; 1990.
-
- Apse M P, Aharon G S, Snedden W A, Blumwald E. Science. 1999;285:1256–1258. - PubMed
-
- Rubio F, Gassmann W, Schroeder J I. Science. 1995;270:1660–1663. - PubMed
-
- Epstein E, Norlyn J D, Rush D W, Kingsbury R W, Kelley D B, Cunningham G A, Wrona A F. Science. 1980;210:399–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

