Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling
- PMID: 10725385
- PMCID: PMC16311
- DOI: 10.1073/pnas.97.7.3747
Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling
Abstract
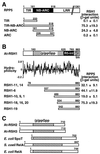
Arabidopsis RPP5 is a member of a large class of pathogen resistance genes encoding nucleotide-binding sites and leucine-rich repeat domains. Yeast two-hybrid analysis showed that RPP5 specifically interacts with At-RSH1, an Arabidopsis RelA/SpoT homolog. In Escherichia coli, RelA and SpoT determine the level of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), which are the effector nucleotides of the bacterial stringent response. Functional analysis in E. coli and in Streptomyces coelicolor A3 (2) showed that At-RSH1 confers phenotypes associated with (p)ppGpp synthesis. We characterized two additional Arabidopsis RelA/SpoT homologs, At-RSH2 and At-RSH3. At-RSH genes may regulate a rapid plant (p)ppGpp-mediated response to pathogens and other stresses.
Figures



Similar articles
-
Plastidial (p)ppGpp Synthesis by the Ca2+-Dependent RelA-SpoT Homolog Regulates the Adaptation of Chloroplast Gene Expression to Darkness in Arabidopsis.Plant Cell Physiol. 2021 Feb 4;61(12):2077-2086. doi: 10.1093/pcp/pcaa124. Plant Cell Physiol. 2021. PMID: 33089303
-
SpoT Induces Intracellular Salmonella Virulence Programs in the Phagosome.mBio. 2020 Feb 25;11(1):e03397-19. doi: 10.1128/mBio.03397-19. mBio. 2020. PMID: 32098823 Free PMC article.
-
Within and beyond the stringent response-RSH and (p)ppGpp in plants.Planta. 2017 Nov;246(5):817-842. doi: 10.1007/s00425-017-2780-y. Epub 2017 Sep 25. Planta. 2017. PMID: 28948393 Free PMC article. Review.
-
Plant RelA/SpoT homolog confers salt tolerance in Escherichia coli and Saccharomyces cerevisiae.Plant Cell Physiol. 2003 Jan;44(1):3-9. doi: 10.1093/pcp/pcg001. Plant Cell Physiol. 2003. PMID: 12552141
-
Recent functional insights into the role of (p)ppGpp in bacterial physiology.Nat Rev Microbiol. 2015 May;13(5):298-309. doi: 10.1038/nrmicro3448. Epub 2015 Apr 8. Nat Rev Microbiol. 2015. PMID: 25853779 Free PMC article. Review.
Cited by
-
The Arabidopsis cold-responsive transcriptome and its regulation by ICE1.Plant Cell. 2005 Nov;17(11):3155-75. doi: 10.1105/tpc.105.035568. Epub 2005 Oct 7. Plant Cell. 2005. PMID: 16214899 Free PMC article.
-
Identification of the bacterial alarmone guanosine 5'-diphosphate 3'-diphosphate (ppGpp) in plants.Proc Natl Acad Sci U S A. 2004 Mar 23;101(12):4320-4. doi: 10.1073/pnas.0308555101. Epub 2004 Mar 9. Proc Natl Acad Sci U S A. 2004. PMID: 15010537 Free PMC article.
-
Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana.Planta. 2008 Sep;228(4):553-62. doi: 10.1007/s00425-008-0758-5. Epub 2008 Jun 6. Planta. 2008. PMID: 18535838
-
Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus.J Bacteriol. 2004 Jan;186(1):235-9. doi: 10.1128/JB.186.1.235-239.2004. J Bacteriol. 2004. PMID: 14679243 Free PMC article.
-
Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription?Plant Mol Biol. 2011 Jul;76(3-5):235-49. doi: 10.1007/s11103-010-9714-4. Epub 2010 Nov 25. Plant Mol Biol. 2011. PMID: 21107995
References
-
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. - PubMed
-
- Galán J E, Collmer A. Science. 1999;284:1322–1328. - PubMed
-
- Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Mol Biol. 1997;48:575–607. - PubMed
-
- Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. - PubMed
-
- Ritter C, Dangl J L. Mol Plant Microbe–Interact. 1995;8:444–453. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

