A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing
- PMID: 10729146
- PMCID: PMC111880
- DOI: 10.1128/jvi.74.8.3709-3714.2000
A novel subgenomic murine leukemia virus RNA transcript results from alternative splicing
Abstract
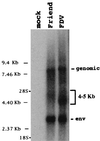
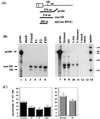
Here we show the existence of a novel subgenomic 4.4-kb RNA in cells infected with the prototypic replication-competent Friend or Moloney murine leukemia viruses (MuLV). This RNA derives by splicing from an alternative donor site (SD') within the capsid-coding region to the canonical envelope splice acceptor site. The position and the sequence of SD' was highly conserved among mammalian type C and D oncoviruses. Point mutations used to inactivate SD' without changing the capsid-coding ability affected viral RNA splicing and reduced viral replication in infected cells.
Figures






References
-
- Barklis E, Mulligan R C, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. - PubMed
-
- Eiden M, Trainor C D, Reitz M S. Gibbon ape leukaemia virus RNA in leukaemic T-lymphoid cell lines: expression of a novel RNA transcript. J Gen Virol. 1986;67:1455–1460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

