mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development
- PMID: 10733527
- PMCID: PMC316460
mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development
Abstract
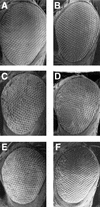
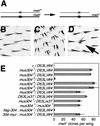
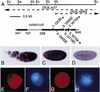
Checkpoints block cell cycle progression in eukaryotic cells exposed to DNA damaging agents. We show that several Drosophila homologs of checkpoint genes, mei-41, grapes, and 14-3-3epsilon, regulate a DNA damage checkpoint in the developing eye. We have used this assay to show that the mutagen-sensitive gene mus304 is also required for this checkpoint. mus304 encodes a novel coiled-coil domain protein, which is targeted to the cytoplasm. Similar to mei-41, mus304 is required for chromosome break repair and for genomic stability. mus304 animals also exhibit three developmental defects, abnormal bristle morphology, decreased meiotic recombination, and arrested embryonic development. We suggest that these phenotypes reflect distinct developmental consequences of a single underlying checkpoint defect. Similar mechanisms may account for the puzzling array of symptoms observed in humans with mutations in the ATM tumor suppressor gene.
Figures




References
-
- Ashburner M. Drosophila: A laboratory handbook. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
