The carboxyl terminus of phage HK022 Nun includes a novel zinc-binding motif and a tryptophan required for transcription termination
- PMID: 10733532
- PMCID: PMC316458
The carboxyl terminus of phage HK022 Nun includes a novel zinc-binding motif and a tryptophan required for transcription termination
Abstract
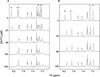
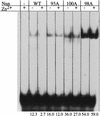
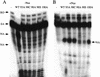
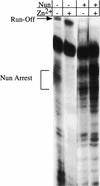
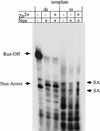
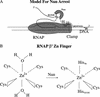
The amino-terminal arginine-rich motif of the phage HK022 Nun protein binds phage lambda nascent mRNA transcripts while the carboxy-terminal domain binds RNA polymerase and arrests transcription. The role of specific residues in the carboxy-terminal domain in transcription termination were investigated by mutagenesis, in vitro and in vivo functional assays, and NMR spectroscopy. Coordination of zinc to three histidine residues in the carboxy-terminus inhibited RNA binding by the amino-terminal domain; however, only two of these histidines were required for transcription arrest. These results suggest that additional zinc-coordinating residues are supplied by RNA polymerase in the context of the Nun-RNA polymerase complex. Substitution of the penultimate carboxy-terminal tryptophan residue with alanine or leucine blocks transcription arrest, whereas a tyrosine substitution is innocuous. Wild-type Nun fails to arrest transcription on single-stranded templates. These results suggest that Nun inhibition of transcription elongation is due in part to interactions between the carboxy-terminal tryptophan of Nun and double-stranded DNA, possibly by intercalation. A model for the termination activity of Nun is developed on the basis of these data.
Figures
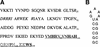
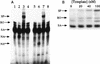
References
-
- Berg J. Zinc finger domains: Hypotheses and current knowledge. Annu Rev of Biophys & Biophys Chem. 1990;19:405–421. - PubMed
-
- Brown R, Sander C, Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Letters. 1985;186:271–274. - PubMed
-
- Burova E, Hung SC, Chen J, Court DL, Zhou J-G, Mogilnitskiy G, Gottesman ME. E. coli mutations that block transcription termination by coliphage HK022 Nun protein. Mol Microbiol. 1999;31:1783–1794. - PubMed
-
- Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: Principles and practice. San Diego, CA: Academic Press; 1996.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
