Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal
- PMID: 10733568
- PMCID: PMC85481
- DOI: 10.1128/MCB.20.8.2660-2669.2000
Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal
Abstract
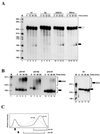
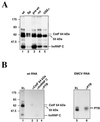
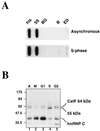
We have investigated how the upstream sequence element (USE) of the lamin B2 poly(A) signal mediates efficient 3'-end formation. In vitro analysis demonstrates that this USE increases both the efficiency of 3'-end cleavage and the processivity of poly(A) addition. Cross-linking using selectively labeled synthetic RNAs confirms that cleavage stimulation factor interacts with the sequences downstream of the cleavage site, while electrophoresis mobility shift assays demonstrate that the USE directly stabilizes the binding of the cleavage and polyadenylation specificity factor to the poly(A) signal. Thus in common with other poly(A) signals, the lamin B2 USE directly enhances the binding of basal poly(A) factors. In addition, a novel 55-kDa protein binds to the USE and the core poly(A) signal and appears to inhibit cleavage. The binding activity of this factor appears to change during the cell cycle, being greatest in S phase, when the lamin B2 gene is transcribed.
Figures




References
-
- Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. - PubMed
-
- Barabino S M, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1706. - PubMed
-
- Bardwell V J, Wickens M, Bienroth S, Keller W, Sproat B S, Lamond A I. Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell. 1991;65:125–133. - PubMed
-
- Biamonti G, Giacca M, Perini G, Contreas G, Zentilin L, Weighardt H, Guerra M, Della Valle G, Saccone S, Riva S, Falaschi A. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S phase. Mol Cell Biol. 1992;12:3499–3506. - PMC - PubMed
-
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing on messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
