Functionally significant secondary structure of the simian virus 40 late polyadenylation signal
- PMID: 10733596
- PMCID: PMC85533
- DOI: 10.1128/MCB.20.8.2926-2932.2000
Functionally significant secondary structure of the simian virus 40 late polyadenylation signal
Abstract
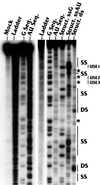
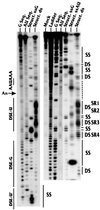
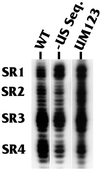
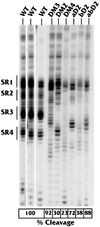
The structure of the highly efficient simian virus 40 late polyadenylation signal (LPA signal) is more complex than those of most known mammalian polyadenylation signals. It contains efficiency elements both upstream and downstream of the AAUAAA region, and the downstream region contains three defined elements (two U-rich elements and one G-rich element) instead of the single U- or GU-rich element found in most polyadenylation signals. Since many reports have indicated that the secondary structure in RNA may play a significant role in RNA processing, we have used nuclease structure analysis techniques to determine the secondary structure of the LPA signal. We find that the LPA signal has a functionally significant secondary structure. Much of the region upstream of AAUAAA is sensitive to single-strand-specific nucleases. The region downstream of AAUAAA has both double- and single-stranded characteristics. Both U-rich elements are predominately sensitive to the double-strand-specific nuclease RNase V(1), while the G-rich element is primarily single stranded. The U-rich element closest to AAUAAA contains four distinct RNase V(1)-sensitive regions, which we have designated structural region 1 (SR1), SR2, SR3, and SR4. Linker scanning mutants in the downstream region were analyzed both for structure and for function by in vitro cleavage analyses. These data show that the ability of the downstream region, particularly SR3, to form double-stranded structures correlates with efficient in vitro cleavage. We discuss the possibility that secondary structure downstream of the AAUAAA may be important for the functions of polyadenylation signals in general.
Figures

References
-
- Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
