Degradation of ribosomal RNA precursors by the exosome
- PMID: 10734186
- PMCID: PMC102825
- DOI: 10.1093/nar/28.8.1684
Degradation of ribosomal RNA precursors by the exosome
Abstract
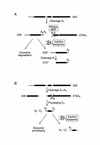
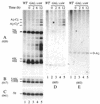
The yeast exosome is a complex of 3'-->5' exonucleases involved in RNA processing and degradation. All 11 known components of the exosome are required during 3' end processing of the 5.8S rRNA. Here we report that depletion of each of the individual components inhibits the early pre-rRNA cleavages at sites A(0), A(1), A(2)and A(3), reducing the levels of the 32S, 20S, 27SA(2)and 27SA(3)pre-rRNAs. The levels of the 27SB pre-rRNAs were also reduced. Consequently, both the 18S and 25S rRNAs were depleted. Since none of these processing steps involves 3'-->5' exonuclease activities, the requirement for the exosome is probably indirect. Correct assembly of trans -acting factors with the pre-ribosomes may be monitored by a quality control system that inhibits pre-rRNA processing. The exosome itself degrades aberrant pre-rRNAs that arise from such inhibition. Exosome mutants stabilize truncated versions of the 23S, 21S and A(2)-C(2)RNAs, none of which are observed in wild-type cells. The putative helicase Dob1p, which functions as a cofactor for the exosome in pre-rRNA processing, also functions in these pre-rRNA degradation activities.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

