Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast
- PMID: 10734188
- PMCID: PMC102823
- DOI: 10.1093/nar/28.8.1700
Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast
Abstract
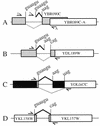
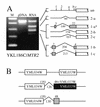
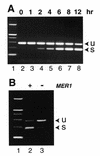
Correct identification of all introns is necessary to discern the protein-coding potential of a eukaryotic genome. The existence of most of the spliceosomal introns predicted in the genome of Saccharomyces cerevisiae remains unsupported by molecular evidence. We tested the intron predictions for 87 introns predicted to be present in non-ribosomal protein genes, more than a third of all known or suspected introns in the yeast genome. Evidence supporting 61 of these predictions was obtained, 20 predicted intron sequences were not spliced and six predictions identified an intron-containing region but failed to specify the correct splice sites, yielding a successful prediction rate of <80%. Alternative splicing has not been previously described for this organism, and we identified two genes (YKL186C/ MTR2 and YML034W) which encode alternatively spliced mRNAs; YKL186C/ MTR2 produces at least five different spliced mRNAs. One gene (YGR225W/ SPO70 ) has an intron whose removal is activated during meiosis under control of the MER1 gene. We found eight new introns, suggesting that numerous introns still remain to be discovered. The results show that correct prediction of introns remains a significant barrier to understanding the structure, function and coding capacity of eukaryotic genomes, even in a supposedly simple system like yeast.
Figures

References
-
- Mewes H.W. et al. (1997) Nature, 387 (suppl.), 7–65. - PubMed
-
- Ainscough R. et al. (1998) Science, 282, 2012–2018. - PubMed
-
- Ansari-Lari M.A., Oeltjen,J.C., Schwartz,S., Zhang,Z., Muzny,D.M., Lu,J., Gorrell,J.H., Chinault,A.C., Belmont,J.W., Miller,W. and Gibbs,R.A. (1998) Genome Res., 8, 29–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

