The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12
- PMID: 10734204
- PMCID: PMC102813
- DOI: 10.1093/nar/28.8.1838
The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12
Abstract
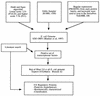
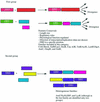
Using a combination of several approaches we estimated and characterized a total of 314 regulatory DNA-binding proteins in Escherichia coli, which might represent its minimal set of transcription factors. The collection is comprised of 35% activators, 43% repressors and 22% dual regulators. Within many regulatory protein families, the members are homogeneous in their regulatory roles, physiology of regulated genes, regulatory function, length and genome position, showing that these families have evolved homogeneously in prokaryotes, particularly in E.coli. This work describes a full characterization of the repertoire of regulatory interactions in a whole living cell. This repertoire should contribute to the interpretation of global gene expression profiles in both prokaryotes and eukaryotes.
Figures

References
-
- Neidhardt F.C., Ingraham,J.L, Low,K.B., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds) (1987) Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC.
-
- Neidhardt F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley.M., Schaerchter,M. and Umbarger,H.E. (eds) (1996) Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. American Society for Microbiology, Washington, DC.
-
- Blattner F.R., Plunkett,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) Science, 277, 1453–1462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

