A novel phenanthrene dioxygenase from Nocardioides sp. Strain KP7: expression in Escherichia coli
- PMID: 10735855
- PMCID: PMC111261
- DOI: 10.1128/JB.182.8.2134-2141.2000
A novel phenanthrene dioxygenase from Nocardioides sp. Strain KP7: expression in Escherichia coli
Abstract
Nocardioides sp. strain KP7 grows on phenanthrene but not on naphthalene. This organism degrades phenanthrene via 1-hydroxy-2-naphthoate, o-phthalate, and protocatechuate. The genes responsible for the degradation of phenanthrene to o-phthalate (phd) were found by Southern hybridization to reside on the chromosome. A 10.6-kb DNA fragment containing eight phd genes was cloned and sequenced. The phdA, phdB, phdC, and phdD genes, which encode the alpha and beta subunits of the oxygenase component, a ferredoxin, and a ferredoxin reductase, respectively, of phenanthrene dioxygenase were identified. The gene cluster, phdAB, was located 8. 3 kb downstream of the previously characterized phdK gene, which encodes 2-carboxybenzaldehyde dehydrogenase. The phdCD gene cluster was located 2.9 kb downstream of the phdB gene. PhdA and PhdB exhibited moderate (less than 60%) sequence identity to the alpha and beta subunits of other ring-hydroxylating dioxygenases. The PhdC sequence showed features of a [3Fe-4S] or [4Fe-4S] type of ferredoxin, not of the [2Fe-2S] type of ferredoxin that has been found in most of the reported ring-hydroxylating dioxygenases. PhdD also showed moderate (less than 40%) sequence identity to known reductases. The phdABCD genes were expressed poorly in Escherichia coli, even when placed under the control of strong promoters. The introduction of a Shine-Dalgarno sequence upstream of each initiation codon of the phdABCD genes improved their expression in E. coli. E. coli cells carrying phdBCD or phdACD exhibited no phenanthrene-degrading activity, and those carrying phdABD or phdABC exhibited phenanthrene-degrading activity which was significantly less than that in cells carrying the phdABCD genes. It was thus concluded that all of the phdABCD genes are necessary for the efficient expression of phenanthrene-degrading activity. The genetic organization of the phd genes, the phylogenetically diverged positions of these genes, and an unusual type of ferredoxin component suggest phenanthrene dioxygenase in Nocardioides sp. strain KP7 to be a new class of aromatic ring-hydroxylating dioxygenases.
Figures

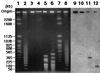
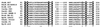

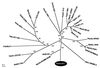
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, Suppl. 11. unit 16.2. New York, N.Y: John Wiley & Sons; 1990.
-
- Batie C J, Ballou D P, Correll C C. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Muller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 3. Boca Raton, Fla: CRC Press; 1992. pp. 543–556.
-
- Bergeron J, Ahmad D, Barriault D, Larose A, Sylvestre M, Powlowski J. Identification and mapping of the gene translation products involved in the first steps of the Comamonas testosteroni B-356 biphenyl/chlorobiphenyl biodegradation pathway. Can J Microbiol. 1994;40:743–753. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

