Recognition of triple-helical DNA structures by transposon Tn7
- PMID: 10737770
- PMCID: PMC18120
- DOI: 10.1073/pnas.080061497
Recognition of triple-helical DNA structures by transposon Tn7
Abstract
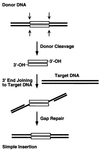
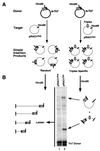
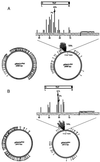
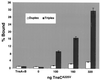
We have found that the bacterial transposon Tn7 can recognize and preferentially insert adjacent to triple-helical nucleic acid structures. Both synthetic intermolecular triplexes, formed through the pairing of a short triplex-forming oligonucleotide on a plasmid DNA, and naturally occurring mirror repeat sequences known to form intramolecular triplexes or H-form DNA are preferential targets for Tn7 insertion in vitro. This target site selectivity depends upon the recognition of the triplex region by a Tn7-encoded ATP-using protein, TnsC, which controls Tn7 target site selection: the interaction of TnsC with the triplex region results in recruitment and activation of the Tn7 transposase. Recognition of a nucleic acid structural motif provides both new information into the factors that influence Tn7's target site selection and broadens its targeting capabilities.
Figures


References
-
- Berg D E, Howe M M, editors. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989.
-
- Craig N L. Annu Rev Biochem. 1997;66:437–474. - PubMed
-
- Berg C M, Berg E. In: Mobile Genetic Elements. Sherratt D J, editor. Oxford: IRL; 1995. pp. 38–68.
-
- Katz R A, Gravuer K, Skalka A M. J Biol Chem. 1998;273:24190–24195. - PubMed
-
- Pryciak P M, Varmus H E. Cell. 1992;69:769–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

