Crystal structure of the functional domain of the splicing factor Prp18
- PMID: 10737784
- PMCID: PMC16185
- DOI: 10.1073/pnas.97.7.3022
Crystal structure of the functional domain of the splicing factor Prp18
Abstract
The splicing factor Prp18 is required for the second step of pre-mRNA splicing. We have isolated and determined the crystal structure of a large fragment of the Saccharomyces cerevisiae Prp18 that lacks the N-terminal 79 amino acids. This fragment, called Prp18Delta79, is fully active in yeast splicing in vitro and includes the sequences of Prp18 that have been evolutionarily conserved. The core structure of Prp18Delta79 is compact and globular, consisting of five alpha-helices that adopt a novel fold that we have designated the five-helix X-bundle. The structure suggests that one face of Prp18 interacts with the splicing factor Slu7, whereas the more evolutionarily conserved amino acids in Prp18 form the opposite face. The most highly conserved region of Prp18, a nearly invariant stretch of 19 aa, forms part of a loop between two alpha-helices and may interact with the U5 small nuclear ribonucleoprotein particles. The structure is consistent with a model in which Prp18 forms a bridge between Slu7 and the U5 small nuclear ribonucleoprotein particles.
Figures
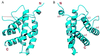



References
-
- Krämer A. Annu Rev Biochem. 1996;65:367–409. - PubMed
-
- Staley J P, Guthrie C. Cell. 1998;92:315–326. - PubMed
-
- Burge C B, Tuschl T H, Sharp P A. In: RNA World II. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560.
-
- Horowitz D S, Abelson J. Genes Dev. 1993;7:320–329. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

