Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase
- PMID: 10737786
- PMCID: PMC16191
- DOI: 10.1073/pnas.97.7.3056
Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase
Abstract
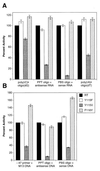
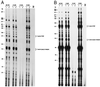
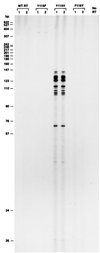
We have examined amino acid substitutions at residues 115 and 116 in the reverse transcriptase (RT) of HIV-1. A number of properties were examined, including polymerization and processivity on both DNA and RNA templates, strand displacement, ribonucleotide misincorporation, and resistance to nucleoside analogs. The RT variants Tyr-115-Phe and Phe-116-Tyr are similar to wild-type HIV-1 RT in most, but not all, respects. In contrast, the RT variant Tyr-115-Val is significantly impaired in polymerase activity compared with wild-type RT; however, Tyr-115-Val is able to incorporate ribonucleotides as well as deoxyribonucleotides during polymerization and is resistant to a variety of nucleoside analogs.
Figures



References
-
- Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. - PubMed
-
- De Clercq E. Biochem Pharmacol. 1994;47:155–169. - PubMed
-
- Mellors J W, Larder B A, Schinazi R G. Int Antiviral News. 1995;3:8–13.
-
- Ding J, Das K, Hsiou Y, Sarafianos S G, Clark A D, Jr, Jacobo-Molina A, Tantillo C, Hughes S H, Arnold E. J Mol Biol. 1998;284:1095–1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

