Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency
- PMID: 10737809
- PMCID: PMC16306
- DOI: 10.1073/pnas.97.7.3718
Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency
Abstract
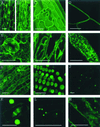
We describe a general approach for identifying components of subcellular structures in a multicellular organism by exploiting the ability to generate thousands of independent transformants in Arabidopsis thaliana. A library of Arabidopsis cDNAs was constructed so that the cDNAs were inserted at the 3' end of the green fluorescent protein (GFP) coding sequence. The library was introduced en masse into Arabidopsis by Agrobacterium-mediated transformation. Fluorescence imaging of 5,700 transgenic plants indicated that approximately 2% of lines expressed a fusion protein with a different subcellular distribution than that of soluble GFP. About half of the markers identified were targeted to peroxisomes or other subcellular destinations by non-native coding sequence (i.e., out-of-frame cDNAs). This observation suggests that some targeting signals are of sufficiently low information content that they can be generated frequently by chance. The potential of the approach for identifying markers with unique dynamic processes is demonstrated by the identification of a GFP fusion protein that displays a cell-cycle regulated change in subcellular distribution. Our results indicate that screening GFP-fusion protein libraries is a useful approach for identifying and visualizing components of subcellular structures and their associated dynamics in higher plant cells.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

