Similar splice-site mutations of the ATP7A gene lead to different phenotypes: classical Menkes disease or occipital horn syndrome
- PMID: 10739752
- PMCID: PMC1288188
- DOI: 10.1086/302857
Similar splice-site mutations of the ATP7A gene lead to different phenotypes: classical Menkes disease or occipital horn syndrome
Abstract
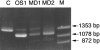
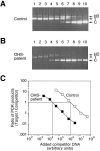
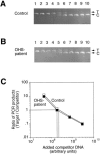
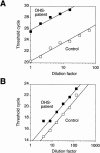
More than 150 point mutations have now been identified in the ATP7A gene. Most of these mutations lead to the classic form of Menkes disease (MD), and a few lead to the milder occipital horn syndrome (OHS). To get a better understanding of molecular changes leading to classic MD and OHS, we took advantage of the unique finding of three patients with similar mutations but different phenotypes. Although all three patients had mutations located in the splice-donor site of intron 6, only two of the patients had the MD phenotype; the third had the OHS phenotype. Fibroblast cultures from the three patients were analyzed by reverse transcriptase (RT)-PCR to try to find an explanation of the different phenotypes. In all three patients, exon 6 was deleted in the majority of the ATP7A transcripts. However, by RT-PCR amplification with an exon 6-specific primer, we were able to amplify exon 6-containing mRNA products from all three patients, even though they were in low abundance. Sequencing of these products indicated that only the patient with OHS had correctly spliced exon 6-containing transcripts. We used two different methods of quantitative RT-PCR analysis and found that the level of correctly spliced mRNA in this patient was 2%-5% of the level found in unaffected individuals. These findings indicate that the presence of barely detectable amounts of correctly spliced ATP7A transcript is sufficient to permit the development of the milder OHS phenotype, as opposed to classic MD.
Figures

References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for MD, MIM 309400) - PubMed
References
-
- Bouaboula M, Legoux P, Pességué B, Delpech B, Dumont X, Piechaczyk M, Casellas P, et al (1992) Standardization of mRNA titration using a polymerase chain reaction method involving co-amplification with a multispecific internal control. J Biol Chem 267:21830–21838 - PubMed
-
- DiDonato M, Sarkar B (1997) Copper transport and its alterations in Menkes and Wilson disease. Biochim Biophys Acta 1360:3–16 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

