Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions
- PMID: 10742240
- PMCID: PMC92021
- DOI: 10.1128/AEM.66.4.1544-1552.2000
Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions
Abstract
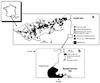
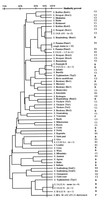
Salmonella species are pathogenic bacteria often detected in sewage, freshwater, marine coastal water, and groundwater. Salmonella spp. can survive for long periods in natural waters, and the persistence of specific and epidemic strains is of great concern in public health. However, the diversity of species found in the natural environment remains unknown. The aim of this study was to investigate the diversity of Salmonella strains isolated from different natural aquatic systems within a Mediterranean coastal watershed (river, wastewater, and marine coastal areas). A total of 574 strains isolated from these natural environments were identified by both conventional serotyping and the ribosomal spacer-heteroduplex polymorphism (RS-HP) method (M. A. Jensen and N. Straus, PCR Methods Appl. 3:186-194, 1993). More than 40 different serotypes were found, and some serotypes probably mobilized from widespread animal-rearing activities were detected only during storm events. These serotypes may be good indicators of specific contamination sources. Furthermore, the RS-HP method based on the PCR amplification of the intergenic spacer region between the 16S and 23S rRNA genes can produce amplicon profiles allowing the discrimination of species at both serotype and intraserotype levels. This method represents a powerful tool that could be used for rapid typing of Salmonella isolates.
Figures


References
-
- Baleux B, Alibou J, Trousselier M, Got P. Utilisation du bouillon Sélénite F modifié pour dénombrer Salmonella dans les milieux aquatiques. Rev Sci Eau. 1988;3:401–408.
-
- Baudart J, Grabulos J, Barusseau J-P, Lebaron P. Salmonella spp. and fecal coliform loads in coastal waters from a point vs. nonpoint source of pollution. J Environ Qual. 2000;29:241–250.
-
- Busse M. Media for Salmonella. Int J Food Microbiol. 1995;26:117–131. - PubMed
-
- Coyne M S, Howell J M. Agricultural impacts on fecal contamination of shallow groundwaters in the Bluegrass region of Kentucky. Soil Sci News Views. 1994;15:1–3.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

