Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter
- PMID: 10742262
- PMCID: PMC92043
- DOI: 10.1128/AEM.66.4.1692-1697.2000
Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter
Abstract
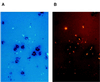
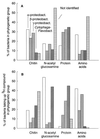
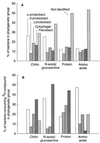
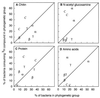
We used a method that combines microautoradiography with hybridization of fluorescent rRNA-targeted oligonucleotide probes to whole cells (MICRO-FISH) to test the hypothesis that the relative contributions of various phylogenetic groups to the utilization of dissolved organic matter (DOM) depend solely on their relative abundance in the bacterial community. We found that utilization of even simple low-molecular-weight DOM components by bacteria differed across the major phylogenetic groups and often did not correlate with the relative abundance of these bacterial groups in estuarine and coastal environments. The Cytophaga-Flavobacter cluster was overrepresented in the portion of the assemblage consuming chitin, N-acetylglucosamine, and protein but was generally underrepresented in the assemblage consuming amino acids. The amino acid-consuming assemblage was usually dominated by the alpha subclass of the class Proteobacteria, although the representation of alpha-proteobacteria in the protein-consuming assemblages was about that expected from their relative abundance in the entire bacterial community. In our experiments, no phylogenetic group dominated the consumption of all DOM, suggesting that the participation of a diverse assemblage of bacteria is essential for the complete degradation of complex DOM in the oceans. These results also suggest that the role of aerobic heterotrophic bacteria in carbon cycling would be more accurately described by using three groups instead of the single bacterial compartment currently used in biogeochemical models.
Figures
References
-
- Amon R M W, Benner R. Bacterial utilization of different size classes of dissolved organic matter. Limnol Oceanogr. 1996;41:41–51.
-
- Amon R M W, Benner R. Rapid cycling of high-molecular-weight dissolved organic matter in the ocean. Nature. 1994;369:549–551.
-
- Barer M R, Entwistle A. Confocal microscopy of surface labeled and cytoplasmically labeled bacteria immobilized by APS centrifugation. Lett Appl Microbiol. 1991;13:186–189.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

