Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors
- PMID: 10744719
- PMCID: PMC2819356
- DOI: 10.1074/jbc.275.14.10315
Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors
Abstract
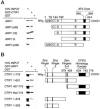
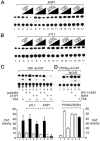
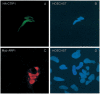
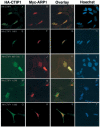
Two novel and related C(2)H(2) zinc finger proteins that are highly expressed in the brain, CTIP1 and CTIP2 (COUP TF-interacting proteins 1 and 2, respectively), were isolated and shown to interact with all members of the chicken ovalbumin upstream promoter transcription factor (COUP-TF) subfamily of orphan nuclear receptors. The interaction of CTIP1 with ARP1 was studied in detail, and CTIP1 was found to harbor two independent ARP1 interaction domains, ID1 and ID2, whereas the putative AF-2 of ARP1 was required for interaction with CTIP1. CTIP1, which exhibited a punctate staining pattern within the nucleus of transfected cells, recruited cotransfected ARP1 to these foci and potentiated ARP1-mediated transcriptional repression of a reporter construct. However, transcriptional repression mediated by ARP1 acting through CTIP1 did not appear to involve recruitment of a trichostatin A-sensitive histone deacetylase(s) to the template, suggesting that this repression pathway may be distinct from that utilized by several other nuclear receptors.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

