DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells
- PMID: 10747040
- PMCID: PMC310241
- DOI: 10.1093/emboj/19.7.1731
DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells
Abstract
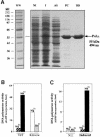
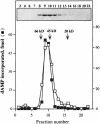
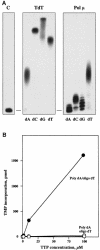
A novel DNA polymerase has been identified in human cells. Human DNA polymerase mu (Pol mu), consisting of 494 amino acids, has 41% identity to terminal deoxynucleotidyltransferase (TdT). Human Pol mu, overproduced in Escherichia coli in a soluble form and purified to homogeneity, displays intrinsic terminal deoxynucleotidyltransferase activity and a strong preference for activating Mn(2+) ions. Interestingly, unlike TdT, the catalytic efficiency of polymerization carried out by Pol mu was enhanced by the presence of a template strand. Using activating Mg(2+) ions, template-enhanced polymerization was also template-directed, leading to the preferred insertion of complementary nucleotides, although with low discrimination values. In the presence of Mn(2+) ions, template-enhanced polymerization produced a random insertion of nucleotides. Northern-blotting and in situ analysis showed a preferential expression of Pol mu mRNA in peripheral lymphoid tissues. Moreover, a large proportion of the human expressed sequence tags corresponding to Pol mu, present in the databases, derived from germinal center B cells. Therefore, Pol mu is a good candidate to be the mutator polymerase responsible for somatic hyper- mutation of immunoglobulin genes.
Figures





References
-
- Baltimore D. (1974) Is terminal deoxynucleotidyl transferase a somatic mutagen in lymphocytes? Nature, 248, 409–411. - PubMed
-
- Bebenek K. and Kunkel, T.A. (1995) Analyzing fidelity of DNA polymerases. Methods Enzymol., 262, 217–232. - PubMed
-
- Beckman R.A., Mildvan, A.S. and Loeb, L.A. (1985) On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry, 24, 5810–5817. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

