Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae
- PMID: 10747092
- PMCID: PMC2175104
- DOI: 10.1083/jcb.149.1.125
Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae
Abstract
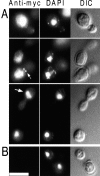
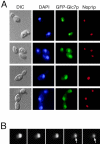
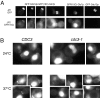
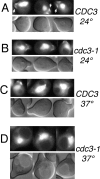
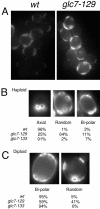
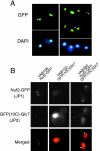
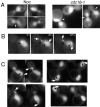
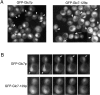
Protein phosphatase type I (PP1), encoded by the single essential gene GLC7 in Saccharomyces cerevisiae, functions in diverse cellular processes. To identify in vivo subcellular location(s) where these processes take place, we used a functional green fluorescent protein (GFP)-Glc7p fusion protein. Time-lapse fluorescence microscopy revealed GFP-Glc7p localizes predominantly in the nucleus throughout the mitotic cell cycle, with the highest concentrations in the nucleolus. GFP-Glc7p was also observed in a ring at the bud neck, which was dependent upon functional septins. Supporting a role for Glc7p in bud site selection, a glc7-129 mutant displayed a random budding pattern. In alpha-factor treated cells, GFP-Glc7p was located at the base of mating projections, again in a septin-dependent manner. At the start of anaphase, GFP-Glc7p accumulated at the spindle pole bodies and remained there until cytokinesis. After anaphase, GFP-Glc7p became concentrated in a ring that colocalized with the actomyosin ring. A GFP-Glc7-129 fusion was defective in localizing to the bud neck and SPBs. Together, these results identify sites of Glc7p function and suggest Glc7p activity is regulated through dynamic changes in its location.
Figures


References
-
- Andrews P.D., Stark M.J. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae . J Cell Sci. 2000;113:507–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

