Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections
- PMID: 10747152
- PMCID: PMC86502
- DOI: 10.1128/JCM.38.4.1615-1622.2000
Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections
Abstract
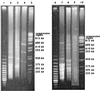
The genetic diversity and relationships among 35 Bacillus cereus and Bacillus thuringiensis isolates recovered from marginal and apical periodontitis in humans and from various other human infections were investigated using multilocus enzyme electrophoresis. The strains were isolated in Norway, except for three strains isolated from periodontitis patients in Brazil. The genetic diversity of these strains was compared to that of 30 isolates from dairies in Norway and Finland. Allelic variation in 13 structural gene loci encoding metabolic enzymes was analyzed. Twelve of the 13 loci were polymorphic, and 48 unique electrophoretic types (ETs) were identified, representing multilocus genotypes. The mean genetic diversity among the 48 genotypes was 0.508. The genetic diversity of each source group of isolates varied from 0.241 (periodontal infection) to 0.534 (dairy). Cluster analysis revealed two major groups separated at a genetic distance of greater than 0.6. One cluster, ETs 1 to 13, included solely isolates from dairies, while the other cluster, ETs 14 to 49, included all of the human isolates as well as isolates from dairies in Norway and Finland. The isolates were serotyped using antiflagellar antiserum. A total of 14 distinct serotypes were observed. However, little association between serotyping and genotyping was seen. Most of the strains were also analyzed with pulsed-field gel electrophoresis, showing the presence of extrachromosomal DNA in the size range of 15 to 600 kb. Our results indicate a high degree of heterogeneity among dairy strains. In contrast, strains isolated from humans had their genotypes in one cluster. Most strains from patients with periodontitis belonged to a single lineage, suggesting that specific clones of B. cereus and B. thuringiensis are associated with oral infections.
Figures

References
-
- Andrup L, Jorgensen O, Wilcks A, Smidt L, Jensen G B. Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid. 1996;36:75–85. - PubMed
-
- Ash C, Farrow J A, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. - PubMed
-
- Carlson C R, Johansen T, M.-M. L, Kolstø A-B. Genomic organization of the entomopathogenic bacterium Bacillus thuringiensis subsp. berliner 1715. Microbiology. 1996;142:1625–1634.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

