A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi
- PMID: 10749927
- PMCID: PMC14844
- DOI: 10.1091/mbc.11.4.1241
A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi
Abstract
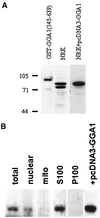
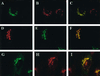
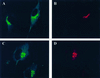
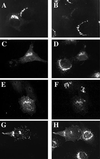
A family of three structurally related proteins were cloned from human cDNA libraries by their ability to interact preferentially with the activated form of human ADP-ribosylation factor 3 (ARF3) in two-hybrid assays. The specific and GTP-dependent binding was later confirmed through direct protein binding of recombinant proteins. The three proteins share large ( approximately 300 residues) domains at their N termini that are 60-70% identical to each other and a shorter (73 residues) domain at their C termini with 70% homology to the C-terminal "ear" domain of gamma-adaptin. Although GGA1 is found predominantly as a soluble protein by cell fractionation, all three proteins were found to localize to the trans-Golgi network (TGN) by indirect immunofluorescence. The binding of GGAs to TGN was sensitive to brefeldin A, consistent with this being an ARF-dependent event. Thus, these proteins have been named Golgi-localizing, gamma-adaptin ear homology domain, ARF-binding proteins, or GGAs. The finding that overexpression of GGAs was sufficient to alter the distribution of markers of the TGN (TGN38 and mannose 6-phosphate receptors) led us to propose that GGAs are effectors for ARFs that function in the regulation of membrane traffic through the TGN.
Figures








References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Balch WE, Kahn RA, Schwaninger R. ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J Biol Chem. 1992;267:13053–13061. - PubMed
-
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
-
- Boman AL, Kahn RA. ARF proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. - PubMed
-
- Boman AL, Taylor TC, Melancon P, Wilson KL. A role for ADP-ribosylation factor in nuclear vesicle dynamics. Nature. 1992;358:512–514. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

