EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans
- PMID: 10749938
- PMCID: PMC14855
- DOI: 10.1091/mbc.11.4.1401
EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans
Abstract
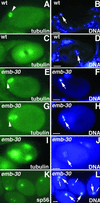
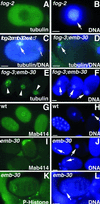
Here we show that emb-30 is required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Germline-specific emb-30 mutant alleles block the meiotic divisions. Mutant oocytes, fertilized by wild-type sperm, set up a meiotic spindle but do not progress to anaphase I. As a result, polar bodies are not produced, pronuclei fail to form, and cytokinesis does not occur. Severe-reduction-of-function emb-30 alleles (class I alleles) result in zygotic sterility and lead to germline and somatic defects that are consistent with an essential role in promoting the metaphase-to-anaphase transition during mitosis. Analysis of the vulval cell lineages in these emb-30(class I) mutant animals suggests that mitosis is lengthened and eventually arrested when maternally contributed emb-30 becomes limiting. By further reducing maternal emb-30 function contributed to class I mutant animals, we show that emb-30 is required for the metaphase-to-anaphase transition in many, if not all, cells. Metaphase arrest in emb-30 mutants is not due to activation of the spindle assembly checkpoint but rather reflects an essential emb-30 requirement for M-phase progression. A reduction in emb-30 activity can suppress the lethality and sterility caused by a null mutation in mdf-1, a component of the spindle assembly checkpoint machinery. This result suggests that delaying anaphase onset can bypass the spindle checkpoint requirement for normal development. Positional cloning established that emb-30 encodes the likely C. elegans orthologue of APC4/Lid1, a component of the anaphase-promoting complex/cyclosome, required for the metaphase-to-anaphase transition. Thus, the anaphase-promoting complex/cyclosome is likely to be required for all metaphase-to-anaphase transitions in a multicellular organism.
Figures







Similar articles
-
Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans.J Cell Biol. 2000 Dec 25;151(7):1469-82. doi: 10.1083/jcb.151.7.1469. J Cell Biol. 2000. PMID: 11134076 Free PMC article.
-
The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans.Curr Biol. 2002 Dec 23;12(24):2118-23. doi: 10.1016/s0960-9822(02)01392-1. Curr Biol. 2002. PMID: 12498686
-
Suppressors of spindle checkpoint defect (such) mutants identify new mdf-1/MAD1 interactors in Caenorhabditis elegans.Genetics. 2007 Apr;175(4):1665-79. doi: 10.1534/genetics.106.067918. Epub 2007 Jan 21. Genetics. 2007. PMID: 17237515 Free PMC article.
-
Control of metaphase-anaphase progression by proteolysis: cyclosome function regulated by the protein kinase A pathway, ubiquitination and localization.Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1559-69; discussion 1569-70. doi: 10.1098/rstb.1999.0499. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582241 Free PMC article. Review.
-
Regulation of APC-Cdc20 by the spindle checkpoint.Curr Opin Cell Biol. 2002 Dec;14(6):706-14. doi: 10.1016/s0955-0674(02)00382-4. Curr Opin Cell Biol. 2002. PMID: 12473343 Review.
Cited by
-
Identification of Conserved MEL-28/ELYS Domains with Essential Roles in Nuclear Assembly and Chromosome Segregation.PLoS Genet. 2016 Jun 24;12(6):e1006131. doi: 10.1371/journal.pgen.1006131. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27341616 Free PMC article.
-
Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I.Genetics. 2002 Feb;160(2):805-13. doi: 10.1093/genetics/160.2.805. Genetics. 2002. PMID: 11861581 Free PMC article.
-
Ubiquitin ligases and a processive proteasome facilitate protein clearance during the oocyte-to-embryo transition in Caenorhabditis elegans.Genetics. 2022 May 5;221(1):iyac051. doi: 10.1093/genetics/iyac051. Genetics. 2022. PMID: 35377419 Free PMC article.
-
The Spindle Assembly Checkpoint Is Not Essential for Viability of Human Cells with Genetically Lowered APC/C Activity.Cell Rep. 2016 Mar 1;14(8):1829-40. doi: 10.1016/j.celrep.2016.01.060. Epub 2016 Feb 18. Cell Rep. 2016. PMID: 26904940 Free PMC article.
-
Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation.Genes Dev. 2004 Jan 1;18(1):88-98. doi: 10.1101/gad.285404. Genes Dev. 2004. PMID: 14724179 Free PMC article.
References
-
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. - PubMed
-
- Albertson DG, Sulston JE, White JG. Cell cycling and DNA replication in a mutant blocked in cell division in the nematode C. elegans. Dev Biol. 1978;63:165–178. - PubMed
-
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. - PubMed
-
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

