Cospeciation and horizontal transmission of avian sarcoma and leukosis virus gag genes in galliform birds
- PMID: 10756010
- PMCID: PMC111912
- DOI: 10.1128/jvi.74.9.3984-3995.2000
Cospeciation and horizontal transmission of avian sarcoma and leukosis virus gag genes in galliform birds
Abstract
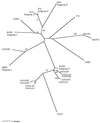
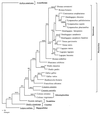
In a study of the evolution and distribution of avian retroviruses, we found avian sarcoma and leukosis virus (ASLV) gag genes in 26 species of galliform birds from North America, Central America, eastern Europe, Asia, and Africa. Nineteen of the 26 host species from whom ASLVs were sequenced were not previously known to contain ASLVs. We assessed congruence between ASLV phylogenies based on a total of 110 gag gene sequences and ASLV-host phylogenies based on mitochondrial 12S ribosomal DNA and ND2 sequences to infer coevolutionary history for ASLVs and their hosts. Widespread distribution of ASLVs among diverse, endemic galliform host species suggests an ancient association. Congruent ASLV and host phylogenies for two species of Perdix, two species of Gallus, and Lagopus lagopus and L. mutus also indicate an old association with vertical transmission and cospeciation for these ASLVs and hosts. An inference of horizontal transmission of ASLVs among some members of the Tetraoninae subfamily (grouse and ptarmigan) is supported by ASLV monophyletic groups reflecting geographic distribution and proximity of hosts rather than host species phylogeny. We provide a preliminary phylogenetic taxonomy for the new ASLVs, in which named taxa denote monophyletic groups.
Figures
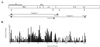



References
-
- Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
-
- Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304.
-
- Cracraft J, Mindell D P. The early history of modern birds: a comparison of molecular and morphological evidence. In: Fernholm B, Bremer K, Jörnvall H, editors. The hierarchy of life: molecules and morphology in phylogenetic analysis. Amsterdam, The Netherlands: Elsevier Science Publishers; 1989. pp. 389–403.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

