Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells
- PMID: 10756014
- PMCID: PMC111916
- DOI: 10.1128/jvi.74.9.4028-4038.2000
Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells
Abstract
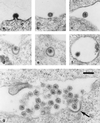
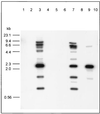
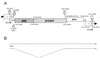
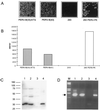
The use of pig xenografts is being considered to alleviate the shortage of allogeneic organs for transplantation. In addition to the problems overcoming immunological and physiological barriers, the existence of numerous porcine microorganisms poses the risk of initiating a xenozoonosis. Recently, different classes of type C porcine endogenous retoviruses (PERV) which are infectious for human cells in vitro have been partially described. We therefore examined whether completely intact proviruses exist that produce infectious and replication-competent virions. Several proviral PERV sequences were cloned and characterized. One molecular PERV class B clone, PERV-B(43), generated infectious particles after transfection into human 293 cells. A second clone, PERV-B(33), which was highly homologous to PERV-B(43), showed a G-to-A mutation in the first start codon (Met to Ile) of the env gene, preventing this provirus from replicating. However, a genetic recombinant, PERV-B(33)/ATG, carrying a restored env start codon, became infectious and could be serially passaged on 293 cells similar to virus clone PERV-B(43). PERV protein expression was detected 24 to 48 h posttransfection (p. t.) using cross-reacting antiserum, and reverse transcriptase activity was found at 12 to 14 days p.t. The transcriptional start and stop sites as well as the splice donor and splice acceptor sites of PERV mRNA were mapped, yielding a subgenomic env transcript of 3. 1 kb. PERV-B(33) and PERV-B(43) differ in the number of copies of a 39-bp segment in the U3 region of the long terminal repeat. Strategies to identify and to specifically suppress or eliminate those proviruses from the pig genome might help in the production of PERV-free animals.
Figures


References
-
- Allan J S. Xenotransplantation at a crossroads: prevention versus progress. Nat Med. 1996;2:18–21. - PubMed
-
- Armstrong J A, Porterfield J S, Madrid A T D. C-type virus particles in pig kidney cell lines. J Gen Virol. 1971;10:195–198. - PubMed
-
- Bach F H, Robson S C, Winkler H, Ferran C, Stuhlmeier K M, Wrighton C J, Hancock W W. Barriers to xenotransplantation. Nat Med. 1995;1:869–873. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

