Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3
- PMID: 10756044
- PMCID: PMC111946
- DOI: 10.1128/jvi.74.9.4291-4301.2000
Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3
Abstract
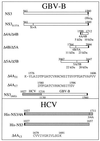
GB virus B (GBV-B) is closely related to hepatitis C virus (HCV) and causes acute hepatitis in tamarins (Saguinus species), making it an attractive surrogate virus for in vivo testing of anti-HCV inhibitors in a small monkey model. It has been reported that the nonstructural protein 3 (NS3) serine protease of GBV-B shares similar substrate specificity with its counterpart in HCV. Authentic proteolytic processing of the HCV polyprotein junctions (NS4A/4B, NS4B/5A, and NS5A/5B) can be accomplished by the GBV-B NS3 protease in an HCV NS4A cofactor-independent fashion. We further characterized the protease activity of a full-length GBV-B NS3 protein and its cofactor requirement using in vitro-translated GBV-B substrates. Cleavages at the NS4A/4B and NS5A/5B junctions were readily detectable only in the presence of a cofactor peptide derived from the central region of GBV-B NS4A. Interestingly, the GBV-B substrates could also be cleaved by the HCV NS3 protease in an HCV NS4A cofactor-dependent manner, supporting the notion that HCV and GBV-B share similar NS3 protease specificity while retaining a virus-specific cofactor requirement. This finding of a strict virus-specific cofactor requirement is consistent with the lack of sequence homology in the NS4A cofactor regions of HCV and GBV-B. The minimum cofactor region that supported GBV-B protease activity was mapped to a central region of GBV-B NS4A (between amino acids Phe22 and Val36) which overlapped with the cofactor region of HCV. Alanine substitution analysis demonstrated that two amino acids, Val27 and Trp31, were essential for the cofactor activity, a finding reminiscent of the two critical residues in the HCV NS4A cofactor, Ile25 and Ile29. A model for the GBV-B NS3 protease domain and NS4A cofactor complex revealed that GBV-B might have developed a similar structural strategy in the activation and regulation of its NS3 protease activity. Finally, a chimeric HCV/GBV-B bifunctional NS3, consisting of an N-terminal HCV protease domain and a C-terminal GBV-B RNA helicase domain, was engineered. Both enzymatic activities were retained by the chimeric protein, which could lead to the development of a chimeric GBV-B virus that depends on HCV protease function.
Figures
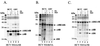
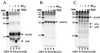




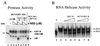

References
-
- Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q-L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A non-B hepatitis. N Engl J Med. 1989;321:1494–1500. - PubMed
-
- Alter M J, Kruszon-Moran D, Nainan O V, McQuillan G M, Gao F, Moyer L A, Kaslow R A, Margolis H S. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. - PubMed
-
- Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

