Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a
- PMID: 10756046
- PMCID: PMC111948
- DOI: 10.1128/jvi.74.9.4310-4318.2000
Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a
Abstract
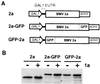
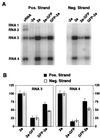
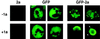
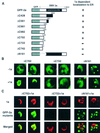
Brome mosaic virus (BMV), a positive-strand RNA virus in the alphavirus-like superfamily, encodes RNA replication proteins 1a and 2a. 1a contains a C-terminal helicase-like domain and an N-terminal domain implicated in viral RNA capping, and 2a contains a central polymerase-like domain. 1a and 2a colocalize in an endoplasmic reticulum (ER)-associated replication complex that is the site of BMV-specific RNA-dependent RNA synthesis in plant and yeast cells. 1a also localizes to the ER in the absence of 2a or viral RNA replication templates. To investigate the determinants of 2a localization, we fused 2a to the green fluorescent protein (GFP), creating a functional GFP-2a fusion that supported BMV RNA replication and subgenomic mRNA transcription. In the absence of 1a, the GFP-2a fusion was found to be diffused throughout the cytoplasm and in punctate spots not associated with any cytoplasmic organelle so far tested. Formation of these spots was dependent on the C-terminal half of 2a and may represent aggregation of a fraction of 2a. When coexpressed with 1a, GFP-2a colocalized with 1a and ER-resident protein Kar2p in a partial or complete ring around the nucleus. Consistent with these results, cell fractionation showed that both the GFP-2a fusion and wild-type (wt) 2a remained soluble when expressed alone, while in cells coexpressing 1a, most of the GFP-2a fusion or wt 2a cofractionated with 1a in the rapidly sedimenting membrane fraction. Deletion analysis showed that the N-terminal 120-amino-acid segment of 2a, containing one of two 2a regions previously shown to interact with 1a, was necessary and sufficient for 1a-directed localization of GFP-2a derivatives to the ER. These results suggest that 1a, which also interacts independently with the ER and viral RNA, is a key organizer of RNA replication complex assembly.
Figures



References
-
- Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

