Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection
- PMID: 10756051
- PMCID: PMC111953
- DOI: 10.1128/jvi.74.9.4361-4376.2000
Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection
Abstract
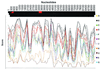
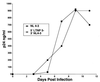
Factors accounting for long-term nonprogression may include infection with an attenuated strain of human immunodeficiency virus type 1 (HIV-1), genetic polymorphisms in the host, and virus-specific immune responses. In this study, we examined eight individuals with nonprogressing or slowly progressing HIV-1 infection, none of whom were homozygous for host-specific polymorphisms (CCR5-Delta32, CCR2-64I, and SDF-1-3'A) which have been associated with slower disease progression. HIV-1 was recovered from seven of the eight, and recovered virus was used for sequencing the full-length HIV-1 genome; full-length HIV-1 genome sequences from the eighth were determined following amplification of viral sequences directly from peripheral blood mononuclear cells (PBMC). Longitudinal studies of one individual with HIV-1 that consistently exhibited a slow/low growth phenotype revealed a single amino acid deletion in a conserved region of the gp41 transmembrane protein that was not seen in any of 131 envelope sequences in the Los Alamos HIV-1 sequence database. Genetic analysis also revealed that five of the eight individuals harbored HIV-1 with unusual 1- or 2-amino-acid deletions in the Gag sequence compared to subgroup B Gag consensus sequences. These deletions in Gag have either never been observed previously or are extremely rare in the database. Three individuals had deletions in Nef, and one had a 4-amino-acid insertion in Vpu. The unusual polymorphisms in Gag, Env, and Nef described here were also found in stored PBMC samples taken 3 to 11 years prior to, or in one case 4 years subsequent to, the time of sampling for the original sequencing. In all, seven of the eight individuals exhibited one or more unusual polymorphisms; a total of 13 unusual polymorphisms were documented in these seven individuals. These polymorphisms may have been present from the time of initial infection or may have appeared in response to immune surveillance or other selective pressures. Our results indicate that unusual, difficult-to-revert polymorphisms in HIV-1 can be found associated with slow progression or nonprogression in a majority of such cases.
Figures












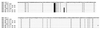




References
-
- Bebenek K, Abbotts J, Roberts J D, Wilson S H, Kunkel T A. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. - PubMed
-
- Binley J M, Jin X, Huang Y, Zhang L, Cao Y, Ho D D, Moore J P. Persistent antibody responses but declining cytotoxic T-lymphocyte responses to multiple human immunodeficiency virus type 1 antigens in a long-term nonprogressing individual with a defective p17 proviral sequence and no detectable viral RNA expression. J Virol. 1998;72:3472–3474. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

