Site-specific modification and RNA crosslinking of the RNA-binding domain of PKR
- PMID: 10756189
- PMCID: PMC103299
- DOI: 10.1093/nar/28.9.1899
Site-specific modification and RNA crosslinking of the RNA-binding domain of PKR
Abstract
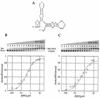
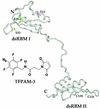
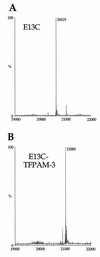
RNA-dependent protein kinase (PKR) is an interferon-induced, RNA-activated enzyme that phosphorylates and inhibits the function of the translation initiation factor eIF-2. PKR is activated in vitro by binding RNA molecules with extensive duplex structure. To further define the nature of the RNA regulation of PKR, we have prepared and characterized site-specifically modified proteins consisting of the PKR 20 kDa RNA-binding domain (RBD). Here we show that the two cysteines found naturally in this domain can be altered by site-directed mutagenesis without loss of RNA binding affinity or the RNA-regulated kinase activity. Introduction of cysteine residues at other sites in the PKR RBD allows for site-specific modification with thiol-selective reagents. PKR RBD mutants reacted selectively with a maleimide to introduce a photoactivatable cross-linking aryl azide at three different positions in the protein. RNA crosslinking efficiency was found to be dependent on the amino acid modified, suggesting differences in access to the RNA from these positions in the protein. One of the amino acid modifications that led to crosslinking of the RNA is located at a residue known to be an autophosphorylation site, suggesting that autophosphorylation at this site could influence the RNA binding properties of PKR. The PKR RBD conjugates described here and other similar reagents prepared via these methods are applicable to future studies of PKR-RNA complexes using techniques such as photocrosslinking, fluorescence resonance energy transfer and affinity cleaving.
Figures



Similar articles
-
Selective binding by the RNA binding domain of PKR revealed by affinity cleavage.Biochemistry. 2001 Apr 10;40(14):4272-80. doi: 10.1021/bi002512w. Biochemistry. 2001. PMID: 11284683
-
A point mutation in the RNA-binding domain I results in decrease of PKR activation in acute lymphoblastic leukemia.Blood Cells Mol Dis. 2005 Jan-Feb;34(1):1-5. doi: 10.1016/j.bcmd.2004.08.025. Blood Cells Mol Dis. 2005. PMID: 15607693
-
trans-Autophosphorylation by the isolated kinase domain is not sufficient for dimerization or activation of the dsRNA-activated protein kinase PKR.Biochemistry. 2004 Aug 31;43(34):11027-34. doi: 10.1021/bi0360105. Biochemistry. 2004. PMID: 15323561
-
Orchestration of the activation of protein kinase R by the RNA-binding motif.J Interferon Cytokine Res. 2010 Apr;30(4):195-204. doi: 10.1089/jir.2010.0005. J Interferon Cytokine Res. 2010. PMID: 20377414 Review.
-
PKR, apoptosis and cancer.Int J Biochem Cell Biol. 1999 Jan;31(1):123-38. doi: 10.1016/s1357-2725(98)00136-8. Int J Biochem Cell Biol. 1999. PMID: 10216948 Review.
Cited by
-
Analysis of PKR activation using analytical ultracentrifugation.Macromol Biosci. 2010 Jul 7;10(7):703-13. doi: 10.1002/mabi.201000069. Macromol Biosci. 2010. PMID: 20533534 Free PMC article.
-
A Nonpolycationic Fully Proteinaceous Multiagent System for Potent Targeted Delivery of siRNA.Mol Ther Nucleic Acids. 2014 May 13;3(5):e162. doi: 10.1038/mtna.2014.14. Mol Ther Nucleic Acids. 2014. PMID: 24825362 Free PMC article.
-
Auto-phosphorylation Represses Protein Kinase R Activity.Sci Rep. 2017 Mar 10;7:44340. doi: 10.1038/srep44340. Sci Rep. 2017. PMID: 28281686 Free PMC article.
-
Phosphorylation of the RNA-dependent protein kinase regulates its RNA-binding activity.Nucleic Acids Res. 2001 Jul 15;29(14):3020-9. doi: 10.1093/nar/29.14.3020. Nucleic Acids Res. 2001. PMID: 11452027 Free PMC article.
-
Cross-linking of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase to template-primer.J Virol. 2001 Oct;75(19):9435-45. doi: 10.1128/JVI.75.19.9435-9445.2001. J Virol. 2001. PMID: 11533206 Free PMC article.
References
-
- Farrell P.J., Balkow,B., Hunt,T. and Jackson,R.J. (1977) Cell, 11, 187–200. - PubMed
-
- Jaramillo M.L., Abraham,N. and Bell,J.C. (1995) Cancer Invest., 13, 327–338. - PubMed
-
- Zhang X., Herring,C.J., Romano,P.R., Szczepanowska,J., Brzeska,H., Hinnebusch,A.G. and Qin,J. (1998) Anal. Chem., 70, 2050–2059. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

