Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5' upstream open reading frames in Fli-1 mRNA
- PMID: 10757781
- PMCID: PMC85554
- DOI: 10.1128/MCB.20.9.2959-2969.2000
Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5' upstream open reading frames in Fli-1 mRNA
Abstract
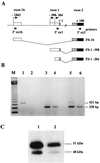
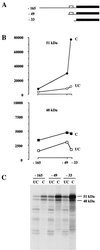
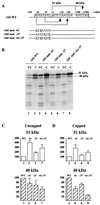
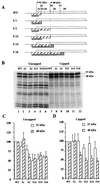
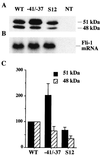
The proto-oncogene Fli-1 encodes a transcription factor of the ets family whose overexpression is associated with multiple virally induced leukemias in mouse, inhibits murine and avian erythroid cell differentiation, and induces drastic perturbations of early development in Xenopus. This study demonstrates the surprisingly sophisticated regulation of Fli-1 mRNA translation. We establish that two FLI-1 protein isoforms (of 51 and 48 kDa) detected by Western blotting in vivo are synthesized by alternative translation initiation through the use of two highly conserved in-frame initiation codons, AUG +1 and AUG +100. Furthermore, we show that the synthesis of these two FLI-1 isoforms is regulated by two short overlapping 5' upstream open reading frames (uORF) beginning at two highly conserved upstream initiation codons, AUG -41 and GUG -37, and terminating at two highly conserved stop codons, UGA +35 and UAA +15. The mutational analysis of these two 5' uORF revealed that each of them negatively regulates FLI-1 protein synthesis by precluding cap-dependent scanning to the 48- and 51-kDa AUG codons. Simultaneously, the translation termination of the two 5' uORF appears to enhance 48-kDa protein synthesis, by allowing downstream reinitiation at the 48-kDa AUG codon, and 51-kDa protein synthesis, by allowing scanning ribosomes to pile up and consequently allowing upstream initiation at the 51-kDa AUG codon. To our knowledge, this is the first example of a cellular mRNA displaying overlapping 5' uORF whose translation termination appears to be involved in the positive control of translation initiation at both downstream and upstream initiation codons.
Figures
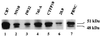




References
-
- Barbeau B, Bergeron D, Beaulieu M, Nadjem Z, Rassart E. Characterization of the human and mouse Fli-1 promoter regions. Biochim Biophys Acta. 1996;1307:220–232. - PubMed
-
- Ben-David Y, Bernstein A. Friend virus induced erythroleukemia and the multistage of cancer. Cell. 1991;66:831–834. - PubMed
-
- Bergeron D, Poliquin L, Houde J, Barbeau B, Rassart E. Analysis of proviruses integrated in Fli-1 and Evi-1 regions in Cas-Br-E MuLV-induced non-T-, non-B-cell leukemias. Virology. 1992;191:661–669. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
