In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs
- PMID: 10757788
- PMCID: PMC85579
- DOI: 10.1128/MCB.20.9.3037-3048.2000
In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs
Abstract
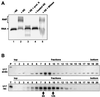
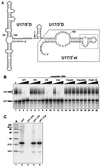
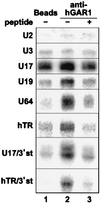
The H/ACA small nucleolar RNAs (snoRNAs) are involved in pseudouridylation of pre-rRNAs. They usually fold into a two-domain hairpin-hinge-hairpin-tail structure, with the conserved motifs H and ACA located in the hinge and tail, respectively. Synthetic RNA transcripts and extracts from HeLa cells were used to reconstitute human U17 and other H/ACA ribonucleoproteins (RNPs) in vitro. Competition and UV cross-linking experiments showed that proteins of about 60, 29, 23, and 14 kDa interact specifically with U17 RNA. Except for U17, RNPs could be reconstituted only with full-length H/ACA snoRNAs. For U17, the 3'-terminal stem-loop followed by box ACA (U17/3'st) was sufficient to form an RNP, and U17/3'st could compete other full-length H/ACA snoRNAs for assembly. The H/ACA-like domain that constitutes the 3' moiety of human telomerase RNA (hTR), and its 3'-terminal stem-loop (hTR/3'st), also could form an RNP by binding H/ACA proteins. Hence, the 3'-terminal stem-loops of U17 and hTR have some specific features that distinguish them from other H/ACA RNAs. Antibodies that specifically recognize the human GAR1 (hGAR1) protein could immunoprecipitate H/ACA snoRNAs and hTR from HeLa cell extracts, which demonstrates that hGAR1 is a component of H/ACA snoRNPs and telomerase in vivo. Moreover, we show that in vitro-reconstituted RNPs contain hGAR1 and that binding of hGAR1 does not appear to be a prerequisite for the assembly of the other H/ACA proteins.
Figures

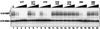



Similar articles
-
Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP.RNA. 2009 Feb;15(2):235-43. doi: 10.1261/rna.1354009. Epub 2008 Dec 17. RNA. 2009. PMID: 19095616 Free PMC article.
-
Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10.Mol Cell Biol. 2000 Dec;20(23):9028-40. doi: 10.1128/MCB.20.23.9028-9040.2000. Mol Cell Biol. 2000. PMID: 11074001 Free PMC article.
-
Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:407-17. doi: 10.1101/sqb.2006.71.025. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381323 Review.
-
hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase.RNA. 2006 May;12(5):832-40. doi: 10.1261/rna.2344106. Epub 2006 Apr 6. RNA. 2006. PMID: 16601202 Free PMC article.
-
The vertebrate E1/U17 small nucleolar ribonucleoprotein particle.J Cell Biochem. 2006 Jun 1;98(3):486-95. doi: 10.1002/jcb.20821. J Cell Biochem. 2006. PMID: 16475166 Review.
Cited by
-
Interaction of Saccharomyces Cdc13p with Pol1p, Imp4p, Sir4p and Zds2p is involved in telomere replication, telomere maintenance and cell growth control.Nucleic Acids Res. 2004 Jan 23;32(2):511-21. doi: 10.1093/nar/gkh203. Print 2004. Nucleic Acids Res. 2004. PMID: 14742666 Free PMC article.
-
Human telomerase and its regulation.Microbiol Mol Biol Rev. 2002 Sep;66(3):407-25, table of contents. doi: 10.1128/MMBR.66.3.407-425.2002. Microbiol Mol Biol Rev. 2002. PMID: 12208997 Free PMC article. Review.
-
Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA.RNA. 2006 Oct;12(10):1868-82. doi: 10.1261/rna.141206. Epub 2006 Aug 24. RNA. 2006. PMID: 16931875 Free PMC article.
-
Naf1 p is a box H/ACA snoRNP assembly factor.RNA. 2002 Dec;8(12):1502-14. RNA. 2002. PMID: 12515383 Free PMC article.
-
Telomere Maintenance and the cGAS-STING Pathway in Cancer.Cells. 2022 Jun 17;11(12):1958. doi: 10.3390/cells11121958. Cells. 2022. PMID: 35741087 Free PMC article. Review.
References
-
- Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem. 1998;273:10868–10873. - PubMed
-
- Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. - PubMed
-
- Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. - PubMed
-
- Blackburn E H. Telomerase. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 609–635.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
