Stress signals utilize multiple pathways to stabilize p53
- PMID: 10757806
- PMCID: PMC85616
- DOI: 10.1128/MCB.20.9.3224-3233.2000
Stress signals utilize multiple pathways to stabilize p53
Abstract
The p53 tumor suppressor is activated by many diverse stress signals through mechanisms that result in stabilization and accumulation of the p53 protein. p53 is normally degraded through the proteasome following interaction with MDM2, which both functions as a ubiquitin ligase for p53 and shuttles to the cytoplasm, where p53 degradation occurs. Stabilization of p53 in response to stress is associated with inhibition of MDM2-mediated degradation, which has been associated with phosphorylation of p53 in response to DNA damage or activation of ARF. In this study we show distinct responses, as measured by phosphorylation, transcriptional activity, and subcellular localization, of p53 stabilized by different activating signals. Although normal cells and wild-type p53-expressing tumor cells showed similar responses to actinomycin D and camptothecin treatment, the transcriptional activity of stabilized p53 induced by deferoxamine mesylate, which mimics hypoxia, in normal cells was lost in all three tumor cell lines tested. Our results show that multiple pathways exist to stabilize p53 in response to different forms of stress, and they may involve down-regulation of MDM2 expression or regulation of the subcellular localization of p53 or MDM2. Loss of any one of these pathways may predispose cells to malignant transformation, although reactivation of p53 might be achieved through alternative pathways that remain functional in these tumor cells.
Figures





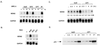



References
-
- An W G, Kanekal M, Simon M C, Maltepe E, Blagosklonny M V, Neckers L M. Stabilization of wild-type by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. - PubMed
-
- Arriola E L, Rodriguez Lopez A, Chresta C M. Differential regulation of p21waf-1/cip-1 and Mdm2 by etoposide: etoposide inhibits the p53-Mdm2 autoregulatory loop. Oncogene. 1999;18:1081–1091. - PubMed
-
- Ashcroft M, Vousden K H. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. - PubMed
-
- Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
