A novel family of cell wall-related proteins regulated differently during the yeast life cycle
- PMID: 10757808
- PMCID: PMC85618
- DOI: 10.1128/MCB.20.9.3245-3255.2000
A novel family of cell wall-related proteins regulated differently during the yeast life cycle
Abstract
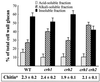
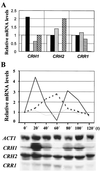
The Saccharomyces cerevisiae Ygr189c, Yel040w, and Ylr213c gene products show significant homologies among themselves and with various bacterial beta-glucanases and eukaryotic endotransglycosidases. Deletion of the corresponding genes, either individually or in combination, did not produce a lethal phenotype. However, the removal of YGR189c and YEL040w, but not YLR213c, caused additive sensitivity to compounds that interfere with cell wall construction, such as Congo red and Calcofluor White, and overexpression of YEL040w led to resistance to these compounds. These genes were renamed CRH1 and CRH2, respectively, for Congo red hypersensitive. By site-directed mutagenesis we found that the putative glycosidase domain of CRH1 was critical for its function in complementing hypersensitivity to the inhibitors. The involvement of CRH1 and CRH2 in the development of cell wall architecture was clearly shown, since the alkali-soluble glucan fraction in the crh1Delta crh2Delta strain was almost twice the level in the wild-type. Interestingly, the three genes were subject to different patterns of transcriptional regulation. CRH1 and YLR213c (renamed CRR1, for CRH related) were found to be cell cycle regulated and also expressed under sporulation conditions, whereas CRH2 expression did not vary during the mitotic cycle. Crh1 and Crh2 are localized at the cell surface, particularly in chitin-rich areas. Consistent with the observed expression patterns, Crh1-green fluorescent protein was found at the incipient bud site, around the septum area in later stages of budding, and in ascospore envelopes. Crh2 was found to localize mainly at the bud neck throughout the whole budding cycle, in mating projections and zygotes, but not in ascospores. These data suggest that the members of this family of putative glycosidases might exert a common role in cell wall organization at different stages of the yeast life cycle.
Figures




References
-
- Arroyo J, Garcia-Gonzalez M, Garcia-Saez M I, Sanchez M, Nombela C. DNA sequence analysis of a 23,002 bp DNA fragment of the right arm of Saccharomyces cerevisiae chromosome VII. Yeast. 1997;13:357–363. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1993.
-
- Cabib E, Roberts R, Bowers B. Synthesis of the yeast cell wall and its regulation. Annu Rev Biochem. 1982;51:763–793. - PubMed
-
- Campbell P, Braam J. Co- and/or post-translational modifications are critical for TCH4 XET activity. Plant J. 1998;15:553–561. - PubMed
-
- Caro L H P, Tettelin H, Vossen J H, Ram A F J, Van Den Ende H, Klis F M. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
