Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression
- PMID: 10757811
- PMCID: PMC85621
- DOI: 10.1128/MCB.20.9.3274-3285.2000
Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression
Abstract
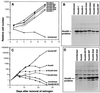
The genes encoding Hoxa9 and Meis1 are transcriptionally coactivated in a subset of acute myeloid leukemia (AML) in mice. In marrow reconstitution experiments, coexpression of both genes produces rapid AML, while neither gene alone generates overt leukemia. Although Hoxa9 and Meis1 can bind DNA as heterodimers, both can also heterodimerize with Pbx proteins. Thus, while their coactivation may result from the necessity to bind promoters as heterodimers, it may also result from the necessity of altering independent biochemical pathways that cooperate to generate AML, either as monomers or as heterodimers with Pbx proteins. Here we demonstrate that constitutive expression of Hoxa9 in primary murine marrow immortalizes a late myelomonocytic progenitor, preventing it from executing terminal differentiation to granulocytes or monocytes in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) or interleukin-3. This immortalized phenotype is achieved in the absence of endogenous or exogenous Meis gene expression. The Hoxa9-immortalized progenitor exhibited a promyelocytic transcriptional profile, expressing PU.1, AML1, c-Myb, C/EBP alpha, and C/EBP epsilon as well as their target genes, the receptors for GM-CSF, G-CSF, and M-CSF and the primary granule proteins myeloperoxidase and neutrophil elastase. G-CSF obviated the differentiation block of Hoxa9, inducing neutrophilic differentiation with accompanying expression of neutrophil gelatinase B and upregulation of gp91phox. M-CSF also obviated the differentiation block, inducing monocytic differentiation with accompanying expression of the macrophage acetyl-low-density lipoprotein scavenger receptor and F4/80 antigen. Versions of Hoxa9 lacking the ANWL Pbx interaction motif (PIM) also immortalized a promyelocytic progenitor with intrinsic biphenotypic differentiation potential. Therefore, Hoxa9 evokes a cytokine-selective block in differentiation by a mechanism that does not require Meis gene expression or interaction with Pbx through the PIM.
Figures






References
-
- Anderson K L, Smith K A, Perkin H, Hermanson G, Anderson C G, Jolly D J, Maki R A, Torbett B E. PU.1 and the granulocyte- and macrophage colony-stimulating factor receptors play distinct roles in late-stage myeloid cell differentiation. Blood. 1999;94:2310–2318. - PubMed
-
- Anderson K L, Smith K A, Pio F, Torbett B E, Maki R A. Neutrophils deficient in PU.1 do not terminally differentiate or become functionally competent. Blood. 1998;92:1576–1585. - PubMed
-
- Austin G E, Zhao W G, Regmi A, Lu J P, Braun J. Identification of an upstream enhancer containing an AML1 site in the human myeloperoxidase (MPO) gene. Leuk Res. 1998;11:1037–1048. - PubMed
-
- Borrow J, Shearman A M, Stanton V P, Jr, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
