Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis
- PMID: 10759555
- PMCID: PMC18185
- DOI: 10.1073/pnas.070430597
Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis
Abstract
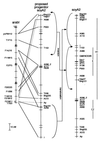
Synteny between soybean and Arabidopsis was studied by using conceptual translations of DNA sequences from loci that map to soybean linkage groups A2, J, and L. Synteny was found between these linkage groups and all four of the Arabidopsis chromosomes, where GenBank contained enough sequence for synteny to be identified confidently. Soybean linkage group A2 (soyA2) and Arabidopsis chromosome I showed significant synteny over almost their entire lengths, with only 2-3 chromosomal rearrangements required to bring the maps into substantial agreement. Smaller blocks of synteny were identified between soyA2 and Arabidopsis chromosomes IV and V (near the RPP5 and RPP8 genes) and between soyA2 and Arabidopsis chromosomes I and V (near the PhyA and PhyC genes). These subchromosomal syntenic regions were themselves homeologous, suggesting that Arabidopsis has undergone a number of segmental duplications or possibly a complete genome duplication during its evolution. Homologies between the homeologous soybean linkage groups J and L and Arabidopsis chromosomes II and IV also revealed evidence of segmental duplication in Arabidopsis. Further support for this hypothesis was provided by the observation of very close linkage in Arabidopsis of homologs of soybean Vsp27 and Bng181 (three locations) and purple acid phosphatase-like sequences and homologs of soybean A256 (five locations). Simulations show that the synteny and duplications we report are unlikely to have arisen by chance during our analysis of the homology reports.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

