A causal link between respiration and senescence in Podospora anserina
- PMID: 10759557
- PMCID: PMC18174
- DOI: 10.1073/pnas.070501997
A causal link between respiration and senescence in Podospora anserina
Abstract
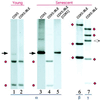
Senescence, a progressive degenerative process leading to age-related increase in mortality, is found in most eukaryotes. However, the molecular events underlying aging remain largely unknown. Understanding how longevity is regulated is a fundamental problem. Here we demonstrate that the respiratory function is a key factor that contributes to shortening lifespan of the filamentous fungus Podospora anserina. In this organism, senescence is systematically associated with mitochondrial DNA instabilities. We show that inactivation of the nuclear COX5 gene encoding subunit V of the cytochrome c oxidase complex leads to the exclusive use of the alternative respiratory pathway and to a decrease in production of reactive oxygen species. This inactivation results in a striking increase of longevity associated with stabilization of the mitochondrial chromosome. Moreover, accumulation of several senescence-specific mitochondrial DNA molecules is prevented in this nuclear mutant. These findings provide direct evidence of a causal link between mitochondrial metabolism and longevity in Podospora anserina.
Figures
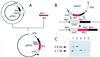


Similar articles
-
Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina.Mol Microbiol. 2001 Dec;42(5):1259-67. doi: 10.1046/j.1365-2958.2001.02690.x. Mol Microbiol. 2001. PMID: 11886557
-
Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina.Biochim Biophys Acta. 2006 May-Jun;1757(5-6):604-10. doi: 10.1016/j.bbabio.2006.03.005. Epub 2006 Mar 30. Biochim Biophys Acta. 2006. PMID: 16624249 Review.
-
Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina.Mol Cell Biol. 2001 Jan;21(2):390-9. doi: 10.1128/MCB.21.2.390-399.2001. Mol Cell Biol. 2001. PMID: 11134328 Free PMC article.
-
Deletion of the mitochondrial NADH kinase increases mitochondrial DNA stability and life span in the filamentous fungus Podospora anserina.Exp Gerontol. 2010 Aug;45(7-8):543-9. doi: 10.1016/j.exger.2010.01.012. Epub 2010 Jan 22. Exp Gerontol. 2010. PMID: 20096769
-
Cell degeneration in the model system Podospora anserina.Biogerontology. 2001;2(1):1-17. doi: 10.1023/a:1010000816277. Biogerontology. 2001. PMID: 11708613 Review.
Cited by
-
Regulation of Aerobic Energy Metabolism in Podospora anserina by Two Paralogous Genes Encoding Structurally Different c-Subunits of ATP Synthase.PLoS Genet. 2016 Jul 21;12(7):e1006161. doi: 10.1371/journal.pgen.1006161. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27442014 Free PMC article.
-
Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts.Antioxid Redox Signal. 2013 Oct 20;19(12):1420-45. doi: 10.1089/ars.2012.5148. Epub 2013 Jul 3. Antioxid Redox Signal. 2013. PMID: 23642158 Free PMC article. Review.
-
Increasing organismal healthspan by enhancing mitochondrial protein quality control.Nat Cell Biol. 2009 Jul;11(7):852-8. doi: 10.1038/ncb1893. Epub 2009 Jun 21. Nat Cell Biol. 2009. PMID: 19543272
-
Lack of mitochondrial citrate synthase discloses a new meiotic checkpoint in a strict aerobe.EMBO J. 2002 Dec 2;21(23):6440-51. doi: 10.1093/emboj/cdf632. EMBO J. 2002. PMID: 12456651 Free PMC article.
-
Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila.Aging (Albany NY). 2010 Apr;2(4):200-23. doi: 10.18632/aging.100137. Aging (Albany NY). 2010. PMID: 20453260 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

