The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor
- PMID: 10760247
- PMCID: PMC139856
- DOI: 10.1105/tpc.12.4.599
The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor
Abstract
The Arabidopsis abscisic acid (ABA)-insensitive abi5 mutants have pleiotropic defects in ABA response, including decreased sensitivity to ABA inhibition of germination and altered expression of some ABA-regulated genes. We isolated the ABI5 gene by using a positional cloning approach and found that it encodes a member of the basic leucine zipper transcription factor family. The previously characterized abi5-1 allele encodes a protein that lacks the DNA binding and dimerization domains required for ABI5 function. Analyses of ABI5 expression provide evidence for ABA regulation, cross-regulation by other ABI genes, and possibly autoregulation. Comparison of seed and ABA-inducible vegetative gene expression in wild-type and abi5-1 plants indicates that ABI5 regulates a subset of late embryogenesis-abundant genes during both developmental stages.
Figures
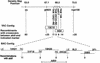
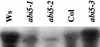




References
-
- Abler, M.L, and Green, P.J.. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32, 63–78. - PubMed
-
- Baeumlein, H., Misera, S., Luerssen, H., Koelle, K., Horstmann, C., Wobus, U., and Mueller, A.J. (1994). The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J. 6, 379–387.
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. - PubMed
-
- Bent, A., Kunkel, B., Dahlbeck, D., Brown, K., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

