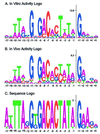
Effects of saturation mutagenesis of the phage SP6 promoter on transcription activity, presented by activity logos
- PMID: 10760261
- PMCID: PMC18112
- DOI: 10.1073/pnas.97.8.3890
Effects of saturation mutagenesis of the phage SP6 promoter on transcription activity, presented by activity logos
Abstract
A full set of SP6 promoter variants with all possible single substitutions at positions -17 to +5 was constructed. Transcription activities of these variants were individually measured in vivo and in vitro to determine the contribution of each base pair to the promoter activity. The in vivo activity was measured indirectly by transcriptional interference of the replication of promoter-bearing plasmids. This activity depends most highly on residues -11, -9, -8, -7, and +1 (initiation site). All substitutions at -11, -9, -8, and -7 abolished formation of closed complexes, except for A-8C. These residues are involved in base-specific interactions with the polymerase, and the substitutions exhibit the same strong inhibition in vitro. In contrast, the in vitro activities of some other variants, measured on linearized templates, were different from those in vivo. Some variants at -13, -4, and -2, among others, showed exceptionally higher activities in vivo than in vitro, supporting the possibility that these residues are involved in postbinding steps, including template melting and bending. The A-3T variant showed much lower activity in vivo than in vitro, but it bound to the polymerase 2-fold more than the consensus sequence and is possibly involved in polymerase binding. A quantitative hierarchy of all the base pairs is graphically displayed by activity logos, revealing the energetic contribution of each base pair to the activity.
Figures


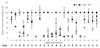


References
-
- Lee S S, Kang C. J Biol Chem. 1992;268:19299–19304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

