Activation of gene expression by small molecule transcription factors
- PMID: 10760265
- PMCID: PMC18119
- DOI: 10.1073/pnas.97.8.3930
Activation of gene expression by small molecule transcription factors
Abstract
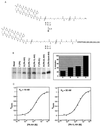
Eukaryotic transcriptional activators are minimally comprised of a DNA binding domain and a separable activation domain; most activator proteins also bear a dimerization module. We have replaced these protein modules with synthetic counterparts to create artificial transcription factors. One of these, at 4.2 kDa, mediates high levels of DNA site-specific transcriptional activation in vitro. This molecule contains a sequence-specific DNA binding polyamide in place of the typical DNA binding region and a nonprotein linker in place of the usual dimerization peptide. Thus our activating region, a designed peptide, functions outside of the archetypal protein context, as long as it is tethered to DNA. Because synthetic polyamides can, in principle, be designed to recognize any specific sequence, these results represent a key step toward the design of small molecules that can up-regulate any specified gene.
Figures


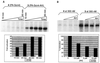
References
-
- Ptashne M, Gann A. Nature (London) 1997;386:569–577. - PubMed
-
- Brent R, Ptashne M. Cell. 1985;43:729–736. - PubMed
-
- Keegan L, Gill G, Ptashne M. Science. 1986;231:699–704. - PubMed
-
- Hope I A, Struhl K. Cell. 1986;46:885–894. - PubMed
-
- Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature (London) 1988;335:563–564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

