Compositional genomes: prebiotic information transfer in mutually catalytic noncovalent assemblies
- PMID: 10760281
- PMCID: PMC18166
- DOI: 10.1073/pnas.97.8.4112
Compositional genomes: prebiotic information transfer in mutually catalytic noncovalent assemblies
Abstract
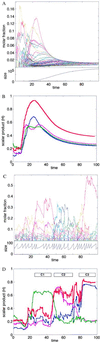
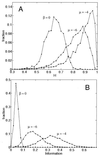
Mutually catalytic sets of simple organic molecules have been suggested to be capable of self-replication and rudimentary chemical evolution. Previous models for the behavior of such sets have analyzed the global properties of short biopolymer ensembles by using graph theory and a mean field approach. In parallel, experimental studies with the autocatalytic formation of amphiphilic assemblies (e.g., lipid vesicles or micelles) demonstrated self-replication properties resembling those of living cells. Combining these approaches, we analyze here the kinetic behavior of small heterogeneous assemblies of spontaneously aggregating molecules, of the type that could form readily under prebiotic conditions. A statistical formalism for mutual rate enhancement is used to numerically simulate the detailed chemical kinetics within such assemblies. We demonstrate that a straightforward set of assumptions about kinetically enhanced recruitment of simple amphiphilic molecules, as well as about the spontaneous growth and splitting of assemblies, results in a complex population behavior. The assemblies manifest a significant degree of homeostasis, resembling the previously predicted quasi-stationary states of biopolymer ensembles (Dyson, F. J. (1982) J. Mol. Evol. 18, 344-350). Such emergent catalysis-driven, compositionally biased entities may be viewed as having rudimentary "compositional genomes." Our analysis addresses the question of how mutually catalytic metabolic networks, devoid of sequence-based biopolymers, could exhibit transfer of chemical information and might undergo selection and evolution. This computed behavior may constitute a demonstration of natural selection in populations of molecules without genetic apparatus, suggesting a pathway from random molecular assemblies to a minimal protocell.
Figures



References
-
- Miller S L. Science. 1953;117:528–529. - PubMed
-
- Hargreaves W R, Mulvihill S, Deamer D W. Nature (London) 1977;266:78–80. - PubMed
-
- Rao M, Eichenberg J, Oró J. J Mol Evol. 1982;18:196–202. - PubMed
-
- Dyson F. Origins of Life. Cambridge: Cambridge Univ. Press; 1999.
-
- Lifson S, Lifson H. J Theor Biol. 1999;199:425–433. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

