Dimers of pi protein bind the A+T-rich region of the R6K gamma origin near the leading-strand synthesis start sites: regulatory implications
- PMID: 10762246
- PMCID: PMC111308
- DOI: 10.1128/JB.182.9.2461-2467.2000
Dimers of pi protein bind the A+T-rich region of the R6K gamma origin near the leading-strand synthesis start sites: regulatory implications
Abstract
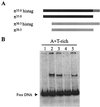
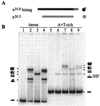
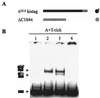
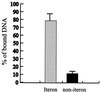
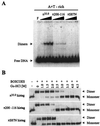
The replication of gamma origin, a minimal replicon derived from plasmid R6K, is controlled by the Rep protein pi. At low intracellular concentrations, pi activates the gamma origin, while it inhibits replication at elevated concentrations. Additionally, pi acts as a transcription factor (auto)repressing its own synthesis. These varied regulatory functions depend on pi binding to reiterated DNA sequences bearing a TGAGNG motif. However, pi also binds to a "non-iteron" site (i.e., not TGAGNG) that resides in the A+T-rich region adjacent to the iterons. This positioning places the non-iteron site near the start sites for leading-strand synthesis that also occur in the A+T-rich region of gamma origin. We have hypothesized that origin activation (at low pi levels) would require the binding of pi monomers to iterons, while the binding of pi dimers to the non-iteron site (at high pi levels) would be required to inhibit priming. Although monomers as well as dimers can bind to an iteron, we demonstrate that only dimers bind to the non-iteron site. Two additional pieces of data support the hypothesis of negative replication control by pi binding to the non-iteron site. First, pi binds to the non-iteron site about eight times less well than it binds to a single iteron. Second, hyperactive variants of pi protein (called copy-up) either do not bind to the non-iteron site or bind to it less well than wild-type pi. We propose a replication control mechanism whereby pi would directly inhibit primer formation.
Figures

References
-
- Bouche J P, Rowen L, Kornberg A. The RNA primer synthesized by primase to initiate phage G4 DNA replication. J Biol Chem. 1978;253:765–769. - PubMed
-
- Bramhill D, Kornberg A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. - PubMed
-
- Chen D, Feng J, Kruger R, Urh M, Inman R B, Filutowicz M. Replication of R6K γ origin in vitro: discrete start sites for DNA synthesis dependent on π and its copy-up variants. J Mol Biol. 1998;282:775–787. - PubMed
-
- Dellis S, Feng J, Filutowicz M. Replication of plasmid R6K γ origin in vivo and in vitro: dependence on IHF binding to the ihf1 site. J Mol Biol. 1996;257:550–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

