Distinct protein interaction domains and protein spreading in a complex centromere
- PMID: 10766735
- PMCID: PMC316498
Distinct protein interaction domains and protein spreading in a complex centromere
Abstract
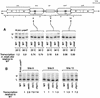
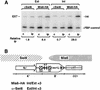
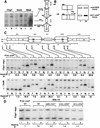
Fission yeast (Schizosaccharomyces pombe) centromeres are composed of large (40-100 kb) inverted repeats that display heterochromatic features, thus providing a good model for higher eukaryotic centromeres. The association of three proteins that mediate region-specific silencing across centromere 1 has been mapped by quantitative chromatin immunoprecipitation. Swi6 and Chp1 are confined to the flanking outer repeats and Swi6 can spread across at least 3 kb of extraneous chromatin in cen1. In contrast, Mis6 coats the inner repeats and central core. tRNA genes demarcate this transition zone. These analyses clearly define two distinct domains within this complex centromere which interact with different proteins.
Figures


References
-
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. - PMC - PubMed
-
- Allshire RC. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. Transcriptional silencing in the fission yeast: A manifestation of higher order chromosome structure and functions; pp. 443–466.
-
- Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. - PubMed
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat J-P, Cranston G. Mutations derepressing silent domains within fission yeast centromeres disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
