Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol
- PMID: 10770760
- PMCID: PMC89853
- DOI: 10.1128/AAC.44.5.1255-1265.2000
Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol
Abstract
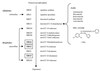
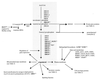
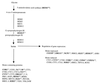
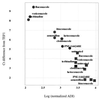
Enzymes in the ergosterol-biosynthetic pathway are the targets of a number of antifungal agents including azoles, allylamines, and morpholines. In order to understand the response of Saccharomyces cerevisiae to perturbations in the ergosterol pathway, genome-wide transcript profiles following exposure to a number of antifungal agents targeting ergosterol biosynthesis (clotrimazole, fluconazole, itraconazole, ketoconazole, voriconazole, terbinafine, and amorolfine) were obtained. These profiles were compared to the transcript profiles of strains containing deletions of one of the late-stage ergosterol genes: ERG2, ERG5, or ERG6. A total of 234 genes were identified as responsive, including the majority of genes from the ergosterol pathway. Expression of several responsive genes, including ERG25, YER067W, and YNL300W, was also monitored by PCR over time following exposure to ketoconazole. The kinetics of transcriptional response support the conditions selected for the microarray experiment. In addition to ergosterol-biosynthetic genes, 36 mitochondrial genes and a number of other genes with roles related to ergosterol function were responsive, as were a number of genes responsive to oxidative stress. Transcriptional changes related to heme biosynthesis were observed in cells treated with chemical agents, suggesting an additional effect of exposure to these compounds. The expression profile in response to a novel imidazole, PNU-144248E, was also determined. The concordance of responsive genes suggests that this compound has the same mode of action as other azoles. Thus, genome-wide transcript profiles can be used to predict the mode of action of a chemical agent as well as to characterize expression changes in response to perturbation of a metabolic pathway.
Figures
References
-
- Amillet J-M, Buisson N, Labbe-Bois R. Characterization of an upstream activation sequence and two Rox1p-responsive sites controlling the induction of the yeast HEM13 gene by oxygen and heme deficiency. J Biol Chem. 1996;271:24425–24432. - PubMed
-
- Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. - PubMed
-
- Boumans H, van Gaalen M C, Grivell L A, Berden J A. Differential inhibition of the yeast bc1 complex by phenanthrolines and ferroin. Implications for structure and catalytic mechanism. J Biol Chem. 1997;272:16753–16760. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

