Differential functional behavior of viral phi29, Nf and GA-1 SSB proteins
- PMID: 10773070
- PMCID: PMC105360
- DOI: 10.1093/nar/28.10.2034
Differential functional behavior of viral phi29, Nf and GA-1 SSB proteins
Abstract
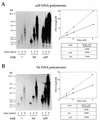
DNA replication of phi29 and related phages takes place via a strand displacement mechanism, a process that generates large amounts of single-stranded DNA (ssDNA). Consequently, phage-encoded ssDNA-binding proteins (SSBs) are essential proteins during phage phi29-like DNA replication. In the present work we analyze the helix-destabilizing activity of the SSBs of phi29 and the related phages Nf and GA-1, their ability to eliminate non-productive binding of phi29 DNA polymerase to ssDNA and their stimulatory effect on replication by phi29 DNA polymerase in primed M13 ssDNA replication, a situation that resembles type II replicative intermediates that occur during phi29-like DNA replication. Significant differences have been appreciated in the functional behavior of the three SSBs. First, the GA-1 SSB is able to display helix-destabilizing activity and to stimulate dNTP incorporation by phi29 DNA polymerase in the M13 DNA replication assay, even at SSB concentrations at which the phi29 and Nf SSBs do not show any effect. On the other hand, the phi29 SSB is the only one of the three SSBs able to increase the replication rate of phi29 DNA polymerase in primed M13 ssDNA replication. From the fact that the phi29 SSB, but not the Nf SSB, stimulates the replication rate of Nf DNA polymerase we conclude that the different behaviors of the SSBs on stimulation of the replication rate of phi29 and Nf DNA polymerases is most likely due to formation of different nucleoprotein complexes of the SSBs with the ssDNA rather than to a specific interaction between the SSB and the corresponding DNA polymerase. A model that correlates the thermodynamic parameters that define SSB-ssDNA nucleoprotein complex formation with the functional stimulatory effect of the SSB on phi29-like DNA replication has been proposed.
Figures





Similar articles
-
DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication.Front Mol Biosci. 2016 Aug 5;3:37. doi: 10.3389/fmolb.2016.00037. eCollection 2016. Front Mol Biosci. 2016. PMID: 27547754 Free PMC article. Review.
-
Strand Displacement and Unwinding Assays to Study the Concerted Action of the DNA Polymerase and SSB During Phi29 TP-DNA Replication.Methods Mol Biol. 2021;2281:333-342. doi: 10.1007/978-1-0716-1290-3_22. Methods Mol Biol. 2021. PMID: 33847970
-
Structural features of phi29 single-stranded DNA-binding protein. II. Global conformation of phi29 single-stranded DNA-binding protein and the effects of complex formation on the protein and the single-stranded DNA.J Biol Chem. 1997 Jan 3;272(1):303-10. doi: 10.1074/jbc.272.1.303. J Biol Chem. 1997. PMID: 8995262
-
Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication.J Mol Biol. 1995 Nov 3;253(4):517-29. doi: 10.1006/jmbi.1995.0570. J Mol Biol. 1995. PMID: 7473731
-
Protein-nucleic acid interactions in bacteriophage phi 29 DNA replication.FEMS Microbiol Rev. 1995 Aug;17(1-2):73-82. doi: 10.1111/j.1574-6976.1995.tb00189.x. FEMS Microbiol Rev. 1995. PMID: 7669351 Review.
Cited by
-
Toward Understanding Phage:Host Interactions in the Rumen; Complete Genome Sequences of Lytic Phages Infecting Rumen Bacteria.Front Microbiol. 2017 Dec 5;8:2340. doi: 10.3389/fmicb.2017.02340. eCollection 2017. Front Microbiol. 2017. PMID: 29259581 Free PMC article.
-
Global Transcriptional Analysis of Virus-Host Interactions between Phage ϕ29 and Bacillus subtilis.J Virol. 2016 Sep 29;90(20):9293-304. doi: 10.1128/JVI.01245-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489274 Free PMC article.
-
Tyrosines involved in the activity of φ29 single-stranded DNA binding protein.PLoS One. 2019 May 20;14(5):e0217248. doi: 10.1371/journal.pone.0217248. eCollection 2019. PLoS One. 2019. PMID: 31107918 Free PMC article.
-
Correction: genomic comparison of 93 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity.BMC Genomics. 2014 Dec 29;15(1):1184. doi: 10.1186/1471-2164-15-1184. BMC Genomics. 2014. PMID: 25547158 Free PMC article.
-
DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication.Front Mol Biosci. 2016 Aug 5;3:37. doi: 10.3389/fmolb.2016.00037. eCollection 2016. Front Mol Biosci. 2016. PMID: 27547754 Free PMC article. Review.
References
-
- Revzin A. (1990) The Biology of Nonspecific DNA–Protein Interactions. CRC Press, Boca Raton, FL.
-
- Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd Edn. Freeman and Co., New York, NY, pp. 323–354.
-
- Chase J.W. and Williams,K.R. (1986) Annu. Rev. Biochem., 55, 103–136. - PubMed
-
- Lohman T.M. and Ferrari,M.E. (1994) Annu. Rev. Biochem., 63, 527–570. - PubMed

