Domain specific interaction in the XRCC1-DNA polymerase beta complex
- PMID: 10773072
- PMCID: PMC105377
- DOI: 10.1093/nar/28.10.2049
Domain specific interaction in the XRCC1-DNA polymerase beta complex
Abstract
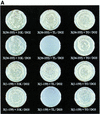
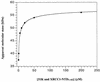
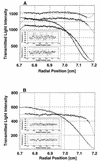
XRCC1 (X-ray cross-complementing group 1) is a DNA repair protein that forms complexes with DNA polymerase beta (beta-Pol), DNA ligase III and poly-ADP-ribose polymerase in the repair of DNA single strand breaks. The domains in XRCC1 have been determined, and characterization of the domain-domain interaction in the XRCC1-beta-Pol complex has provided information on the specificity and mechanism of binding. The domain structure of XRCC1, determined using limited proteolysis, was found to include an N-terminal domain (NTD), a central BRCT-I (breast cancer susceptibility protein-1) domain and a C-terminal BRCT-II domain. The BRCT-I-linker-BRCT-II C-terminal fragment and the linker-BRCT-II C-terminal fragment were relatively stable to proteolysis suggestive of a non-random conformation of the linker. A predicted inner domain was found not to be stable to proteolysis. Using cross-linking experiments, XRCC1 was found to bind intact beta-Pol and the beta-Pol 31 kDa domain. The XRCC1-NTD(1-183)(residues 1-183) was found to bind beta-Pol, the beta-Pol 31 kDa domain and the beta-Pol C-terminal palm-thumb (residues 140-335), and the interaction was further localized to XRCC1-NTD(1-157)(residues 1-157). The XRCC1-NTD(1-183)-beta-Pol 31 kDa domain complex was stable at high salt (1 M NaCl) indicative of a hydrophobic contribution. Using a yeast two-hybrid screen, polypeptides expressed from two XRCC1 constructs, which included residues 36-355 and residues 1-159, were found to interact with beta-Pol, the beta-Pol 31 kDa domain, and the beta-Pol C-terminal thumb-only domain polypeptides expressed from the respective beta-Pol constructs. Neither the XRCC1-NTD(1-159), nor the XRCC1(36-355)polypeptide was found to interact with a beta-Pol thumbless polypeptide. A third XRCC1 polypeptide (residues 75-212) showed no interaction with beta-Pol. In quantitative gel filtration and analytical ultracentrifugation experiments, the XRCC1-NTD(1-183)was found to bind beta-Pol and its 31 kDa domain in a 1:1 complex with high affinity (K(d) of 0.4-2.4 microM). The combined results indicate a thumb-domain specific 1:1 interaction between the XRCC1-NTD(1-159)and beta-Pol that is of an affinity comparable to other binding interactions involving beta-Pol.
Figures


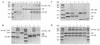

Similar articles
-
Site-directed mutagenesis analysis of the structural interaction of the single-strand-break repair protein, X-ray cross-complementing group 1, with DNA polymerase beta.Nucleic Acids Res. 2003 Jan 15;31(2):580-8. doi: 10.1093/nar/gkg159. Nucleic Acids Res. 2003. PMID: 12527765 Free PMC article.
-
Mapping of the interaction interface of DNA polymerase beta with XRCC1.Structure. 2002 Dec;10(12):1709-20. doi: 10.1016/s0969-2126(02)00908-5. Structure. 2002. PMID: 12467578
-
Solution structure of the single-strand break repair protein XRCC1 N-terminal domain.Nat Struct Biol. 1999 Sep;6(9):884-93. doi: 10.1038/12347. Nat Struct Biol. 1999. PMID: 10467102
-
The structural basis of XRCC1-mediated DNA repair.DNA Repair (Amst). 2015 Jun;30:90-103. doi: 10.1016/j.dnarep.2015.02.005. Epub 2015 Feb 16. DNA Repair (Amst). 2015. PMID: 25795425 Free PMC article. Review.
-
Completion of base excision repair by mammalian DNA ligases.Prog Nucleic Acid Res Mol Biol. 2001;68:151-64. doi: 10.1016/s0079-6603(01)68097-8. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554294 Review.
Cited by
-
BMN673 Is a PARP Inhibitor with Unique Radiosensitizing Properties: Mechanisms and Potential in Radiation Therapy.Cancers (Basel). 2022 Nov 16;14(22):5619. doi: 10.3390/cancers14225619. Cancers (Basel). 2022. PMID: 36428712 Free PMC article. Review.
-
Polβ/XRCC1 heterodimerization dictates DNA damage recognition and basal Polβ protein levels without interfering with mouse viability or fertility.DNA Repair (Amst). 2023 Mar;123:103452. doi: 10.1016/j.dnarep.2023.103452. Epub 2023 Jan 20. DNA Repair (Amst). 2023. PMID: 36702010 Free PMC article.
-
Damage response of XRCC1 at sites of DNA single strand breaks is regulated by phosphorylation and ubiquitylation after degradation of poly(ADP-ribose).J Cell Sci. 2013 Oct 1;126(Pt 19):4414-23. doi: 10.1242/jcs.128272. Epub 2013 Jul 18. J Cell Sci. 2013. PMID: 23868975 Free PMC article.
-
Genetic polymorphisms in the DNA repair genes XPD and XRCC1, p53 gene mutations and bladder cancer risk.Oncol Rep. 2010 Jul;24(1):257-62. doi: 10.3892/or_00000854. Oncol Rep. 2010. PMID: 20514470 Free PMC article.
-
Genetic polymorphisms in XRCC1 associated with radiation therapy in prostate cancer.Cancer Biol Ther. 2010 Jul 1;10(1):13-8. doi: 10.4161/cbt.10.1.12172. Cancer Biol Ther. 2010. PMID: 20495366 Free PMC article.
References
-
- Tebbs R.S., Flannery,M.L., Meneses,J.J., Hartmann,A., Tucker,J.D., Thompson,L.H., Cleaver,J.E. and Pedersen,R.A. (1999) Dev. Biol., 208, 513–529. - PubMed
-
- Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Nature, 379, 183–186. - PubMed
-
- Thompson L.H. and West,M.G. (2000) Mutat. Res., 459, 1–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

