Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA
- PMID: 10773075
- PMCID: PMC105375
- DOI: 10.1093/nar/28.10.2075
Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA
Abstract
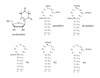
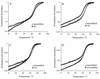
The synthesis of a 5'-O-BzH-2'- O -ACE-protected pseudouridine phosphoramidite is reported [BzH, benzhydryloxy-bis(trimethylsilyloxy)silyl; ACE, bis(2-acetoxyethoxy)methyl]. The availability of the phosphoramidite allows for reliable and efficient syntheses of hairpin RNAs containing single or multiple pseudouridine modifications in the stem or loop regions. Five 19-nt hairpin RNAs representing the 1920-loop region (G(1906)-C(1924)) of Escherichia coli 23S rRNA were synthesized with pseudouridine residues located at positions 1911, 1915 and 1917. Thermodynamic parameters, circular dichroism spectra and NMR data are presented for all five RNAs. Overall, three different structural contexts for the pseudouridine residues were examined and compared with the unmodified RNA. Our main findings are that pseudouridine modifications exhibit a range of effects on RNA stability and structure, depending on their locations. More specifically, pseudouridines in the single-stranded loop regions of the model RNAs are slightly destabilizing, whereas a pseudo-uridine at the stem-loop junction is stabilizing. Furthermore, the observed effects on stability are approximately additive when multiple pseudouridine residues are present. The possible relationship of these results to RNA function is discussed.
Figures



Similar articles
-
Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m(3)Psi and Psi.J Org Chem. 2002 Dec 13;67(25):8847-54. doi: 10.1021/jo026364m. J Org Chem. 2002. PMID: 12467398
-
Synthesis of a 3-methyluridine phosphoramidite to investigate the role of methylation in a ribosomal RNA hairpin.Bioorg Med Chem. 2002 Feb;10(2):325-32. doi: 10.1016/s0968-0896(01)00283-8. Bioorg Med Chem. 2002. PMID: 11741781
-
Pseudouridines in rRNA helix 69 play a role in loop stacking interactions.Org Biomol Chem. 2008 Nov 7;6(21):3892-5. doi: 10.1039/b812731j. Epub 2008 Sep 19. Org Biomol Chem. 2008. PMID: 18931791
-
Pseudouridine in RNA: what, where, how, and why.IUBMB Life. 2000 May;49(5):341-51. doi: 10.1080/152165400410182. IUBMB Life. 2000. PMID: 10902565 Review.
-
The pseudouridine residues of ribosomal RNA.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):915-24. doi: 10.1139/o95-099. Biochem Cell Biol. 1995. PMID: 8722007 Review.
Cited by
-
An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies.Nucleic Acids Res. 2015 Aug 18;43(14):6714-29. doi: 10.1093/nar/gkv606. Epub 2015 Jun 27. Nucleic Acids Res. 2015. PMID: 26117545 Free PMC article.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
-
Investigation of Overhauser effects between pseudouridine and water protons in RNA helices.Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12697-702. doi: 10.1073/pnas.202477199. Epub 2002 Sep 19. Proc Natl Acad Sci U S A. 2002. PMID: 12242344 Free PMC article.
-
SEISMICgraph: a web-based tool for RNA structure data visualization.bioRxiv [Preprint]. 2024 Sep 26:2024.09.26.615187. doi: 10.1101/2024.09.26.615187. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jul 19;53(14):gkaf701. doi: 10.1093/nar/gkaf701. PMID: 39386640 Free PMC article. Updated. Preprint.
-
Site-Specific Synthesis of N4-Acetylcytidine in RNA Reveals Physiological Duplex Stabilization.J Am Chem Soc. 2022 Mar 2;144(8):3487-3496. doi: 10.1021/jacs.1c11985. Epub 2022 Feb 16. J Am Chem Soc. 2022. PMID: 35172571 Free PMC article.
References
-
- Noller H.F. (1999) In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 197–220.
-
- Agris P.F. (1996) Prog. Nucleic Acids Res. Mol. Biol., 53, 79–129. - PubMed
-
- Maden B.E.H. (1990) Prog. Nucleic Acids Res. Mol. Biol., 39, 241–303. - PubMed
-
- Ofengand J. and Bakin,A. (1997) J. Mol. Biol., 266, 246–268. - PubMed
-
- Brimacombe R., Mitchell,P., Osswald,M., Stade,K. and Bochkariov,D. (1993) FASEB J., 7, 161–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

